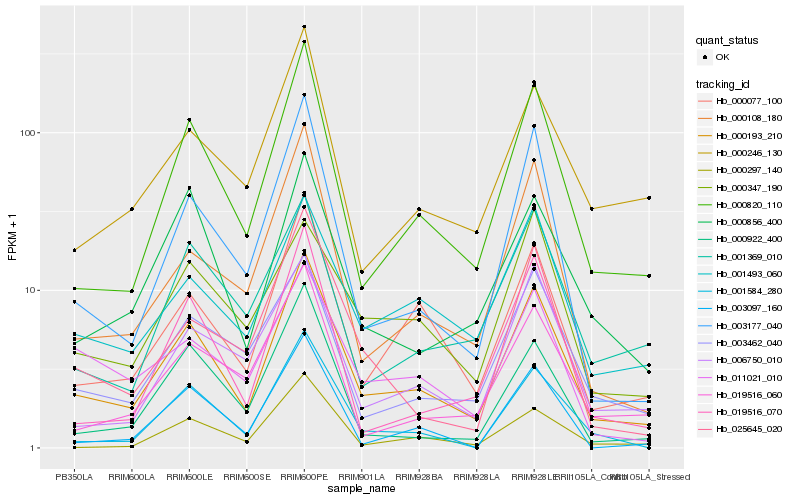
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000193_210 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 2 |
Hb_000820_110 |
0.0929900205 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 3 |
Hb_000077_100 |
0.1219880835 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003177_040 |
0.1292055705 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 5 |
Hb_001493_060 |
0.1386828171 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 6 |
Hb_000108_180 |
0.1395517197 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 7 |
Hb_003462_040 |
0.1425551125 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001584_280 |
0.1433940267 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 9 |
Hb_019516_070 |
0.1451427785 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 10 |
Hb_000856_400 |
0.1458843143 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 11 |
Hb_006750_010 |
0.1471890738 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 12 |
Hb_025645_020 |
0.1504168122 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_000246_130 |
0.1527994249 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 14 |
Hb_000297_140 |
0.1531128883 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 15 |
Hb_019516_060 |
0.1534699096 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 16 |
Hb_001369_010 |
0.1555033701 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 17 |
Hb_000922_400 |
0.1578341201 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 18 |
Hb_011021_010 |
0.158156605 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_003097_160 |
0.1637750843 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 20 |
Hb_000347_190 |
0.1644129938 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |