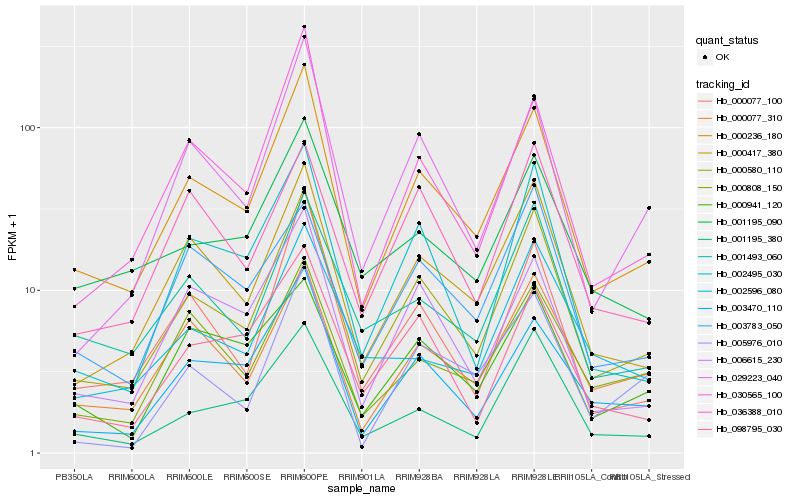
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000580_110 |
0.0 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 2 |
Hb_000236_180 |
0.0661777294 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000417_380 |
0.0902681201 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |
| 4 |
Hb_002495_030 |
0.1101858011 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000077_310 |
0.1148721865 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 6 |
Hb_001195_380 |
0.1156179158 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 7 |
Hb_001493_060 |
0.1158474449 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 8 |
Hb_000077_100 |
0.12340778 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003470_110 |
0.124270605 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_029223_040 |
0.1268441073 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 11 |
Hb_036388_010 |
0.1277100938 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 12 |
Hb_006615_230 |
0.1339796968 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 13 |
Hb_098795_030 |
0.134479159 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 14 |
Hb_000808_150 |
0.1360251522 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 15 |
Hb_030565_100 |
0.1374312805 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 16 |
Hb_001195_090 |
0.1382985596 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 17 |
Hb_003783_050 |
0.1388462273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002596_080 |
0.1408883658 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 19 |
Hb_000941_120 |
0.1432738974 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 20 |
Hb_005976_010 |
0.1437429641 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |