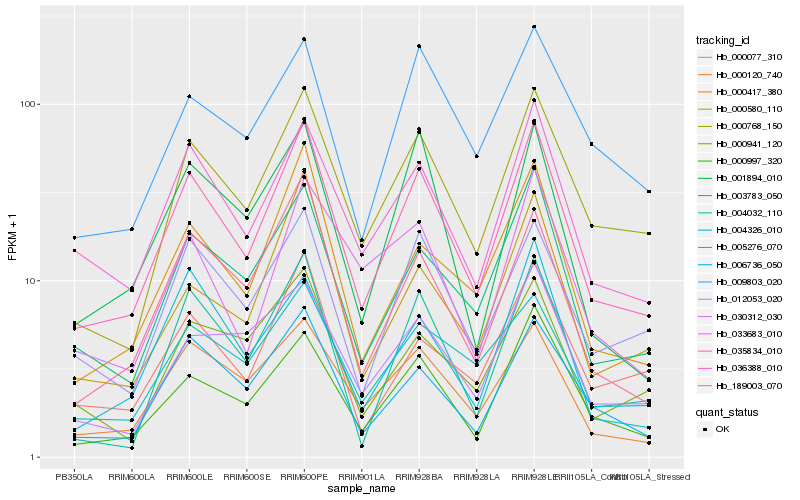
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036388_010 |
0.0 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 2 |
Hb_000768_150 |
0.0915511405 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 3 |
Hb_035834_010 |
0.0934592456 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_003783_050 |
0.0954300344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000997_320 |
0.1003923384 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 6 |
Hb_006736_050 |
0.1031660044 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 7 |
Hb_000417_380 |
0.1059035519 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |
| 8 |
Hb_000120_740 |
0.109430247 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_189003_070 |
0.1101449894 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 10 |
Hb_012053_020 |
0.1144912703 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004032_110 |
0.1199109459 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004326_010 |
0.1214360278 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000077_310 |
0.1243857609 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 14 |
Hb_000580_110 |
0.1277100938 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 15 |
Hb_030312_030 |
0.1282868654 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001894_010 |
0.129602671 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 17 |
Hb_009803_020 |
0.1296788114 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 18 |
Hb_033683_010 |
0.1305459941 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 19 |
Hb_000941_120 |
0.1312483005 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 20 |
Hb_005276_070 |
0.1389921347 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |