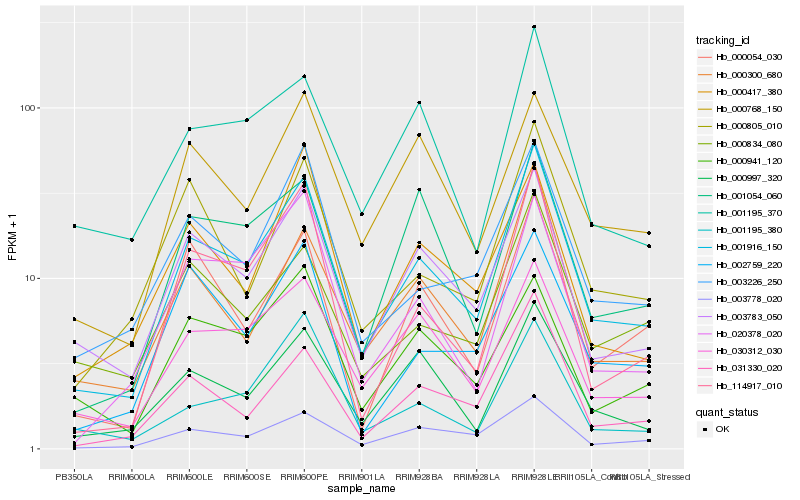
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001916_150 |
0.0 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 2 |
Hb_003783_050 |
0.1031006478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003778_020 |
0.1112600216 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 4 |
Hb_000300_680 |
0.1149545256 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003226_250 |
0.1213814453 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 6 |
Hb_030312_030 |
0.1215082761 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 7 |
Hb_114917_010 |
0.1215146271 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_031330_020 |
0.1240282196 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 9 |
Hb_000997_320 |
0.126689615 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 10 |
Hb_000805_010 |
0.1365361039 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 11 |
Hb_020378_020 |
0.1378197006 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000054_030 |
0.1383856583 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 13 |
Hb_000834_080 |
0.1386136684 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 14 |
Hb_001195_370 |
0.1401755335 |
- |
- |
- |
| 15 |
Hb_001054_060 |
0.1421527643 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 16 |
Hb_002759_220 |
0.1430574061 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 17 |
Hb_000768_150 |
0.1482804217 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 18 |
Hb_000941_120 |
0.1485417758 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 19 |
Hb_001195_380 |
0.1487128739 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 20 |
Hb_000417_380 |
0.1491840333 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |