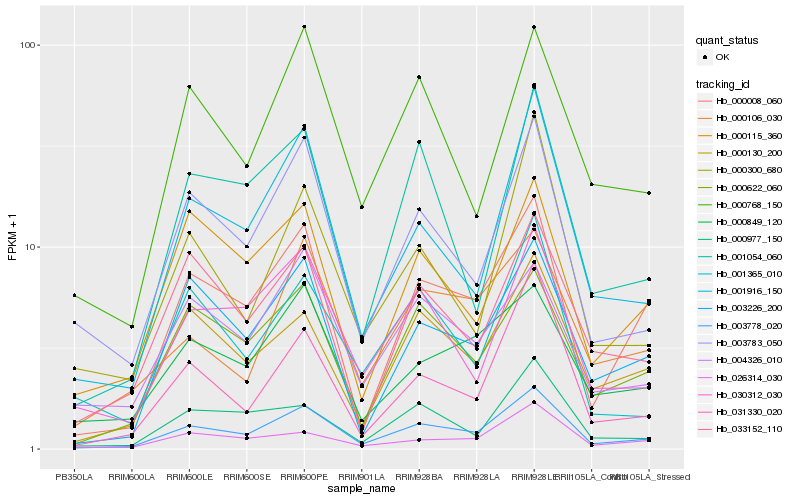
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003778_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 2 |
Hb_000008_060 |
0.109455205 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_001916_150 |
0.1112600216 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 4 |
Hb_003783_050 |
0.1156567088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000130_200 |
0.12219629 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 6 |
Hb_031330_020 |
0.124794693 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 7 |
Hb_000622_060 |
0.1261561983 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003226_200 |
0.1279392787 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 9 |
Hb_030312_030 |
0.1316480182 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 10 |
Hb_026314_030 |
0.1401000625 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 11 |
Hb_033152_110 |
0.1403176436 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 12 |
Hb_000106_030 |
0.142788293 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000977_150 |
0.1432639503 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 14 |
Hb_000768_150 |
0.1433416533 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 15 |
Hb_000849_120 |
0.1433910208 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 16 |
Hb_001054_060 |
0.1436002321 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 17 |
Hb_000115_360 |
0.1438821774 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 18 |
Hb_004326_010 |
0.1457411711 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_000300_680 |
0.1477162624 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001365_010 |
0.1486794671 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |