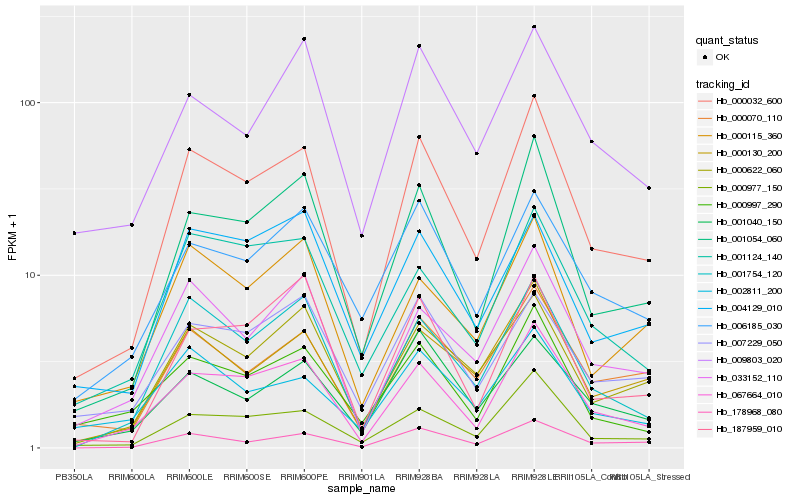
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_600 |
0.0 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 2 |
Hb_067664_010 |
0.0804184791 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 3 |
Hb_001054_060 |
0.0815703156 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 4 |
Hb_000977_150 |
0.0919513586 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 5 |
Hb_178968_080 |
0.0984157513 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000130_200 |
0.1006393522 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 7 |
Hb_001754_120 |
0.1060843884 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 8 |
Hb_000622_060 |
0.1094622234 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_033152_110 |
0.1095278879 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 10 |
Hb_000070_110 |
0.1097852661 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000115_360 |
0.1275770319 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 12 |
Hb_001124_140 |
0.1326423832 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 13 |
Hb_000997_290 |
0.1327651869 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 14 |
Hb_001040_150 |
0.1334113577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_187959_010 |
0.1334532392 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 16 |
Hb_007229_050 |
0.1344678703 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006185_030 |
0.1368399197 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 18 |
Hb_009803_020 |
0.1388118586 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 19 |
Hb_002811_200 |
0.13919054 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 20 |
Hb_004129_010 |
0.1414125994 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |