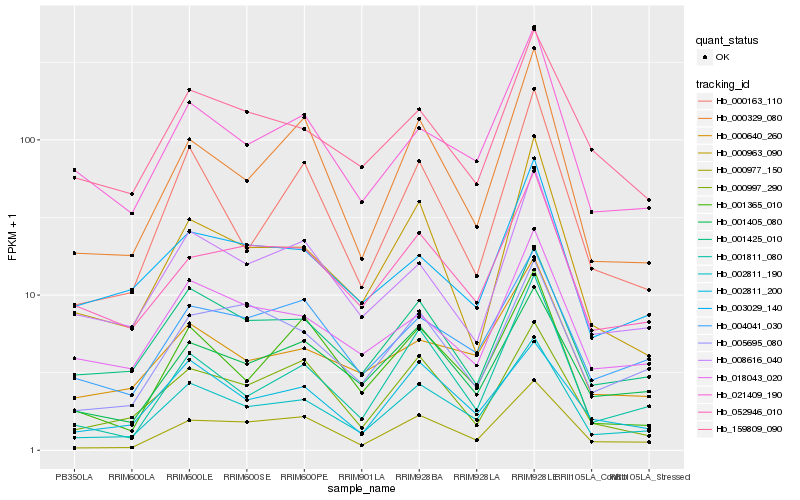
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_190 |
0.0 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 2 |
Hb_001425_010 |
0.09846697 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_000977_150 |
0.1025424513 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 4 |
Hb_001405_080 |
0.1051165732 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 5 |
Hb_021409_190 |
0.1072880488 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 6 |
Hb_000640_260 |
0.1105482192 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000329_080 |
0.1111500799 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004041_030 |
0.1148772587 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 9 |
Hb_002811_200 |
0.1150085352 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 10 |
Hb_001811_080 |
0.1153625067 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 11 |
Hb_052946_010 |
0.1169476473 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 12 |
Hb_000163_110 |
0.1185891796 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 13 |
Hb_018043_020 |
0.1196598655 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_008616_040 |
0.1205613185 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000963_090 |
0.1213713145 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 16 |
Hb_000997_290 |
0.1234701588 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 17 |
Hb_005695_080 |
0.1250578008 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 18 |
Hb_001365_010 |
0.1255697787 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 19 |
Hb_159809_090 |
0.1294176935 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 20 |
Hb_003029_140 |
0.1298828059 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |