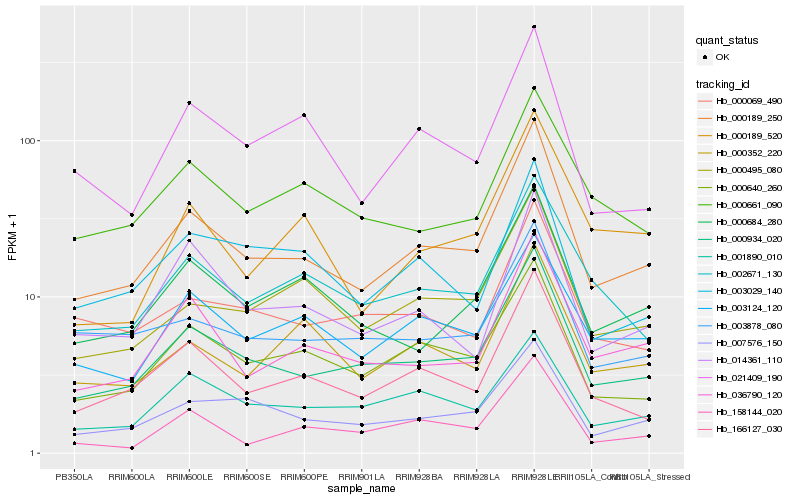
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_250 |
0.0 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000934_020 |
0.0614520208 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 3 |
Hb_166127_030 |
0.0928721696 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000640_260 |
0.1005134099 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 5 |
Hb_036790_120 |
0.1008921242 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007576_150 |
0.1043858661 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g57430, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000495_080 |
0.106663795 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 8 |
Hb_000684_280 |
0.1148397283 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002671_130 |
0.1201114763 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 10 |
Hb_003878_080 |
0.120649767 |
- |
- |
PREDICTED: uncharacterized protein LOC105637045 [Jatropha curcas] |
| 11 |
Hb_014361_110 |
0.1214631798 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000352_220 |
0.1222802368 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000069_490 |
0.1240010341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_021409_190 |
0.1243228167 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 15 |
Hb_001890_010 |
0.1251292829 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 16 |
Hb_003124_120 |
0.1251338875 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 17 |
Hb_000661_090 |
0.1278956419 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 18 |
Hb_158144_020 |
0.1280132123 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 19 |
Hb_000189_520 |
0.1327542277 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 20 |
Hb_003029_140 |
0.1345450434 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |