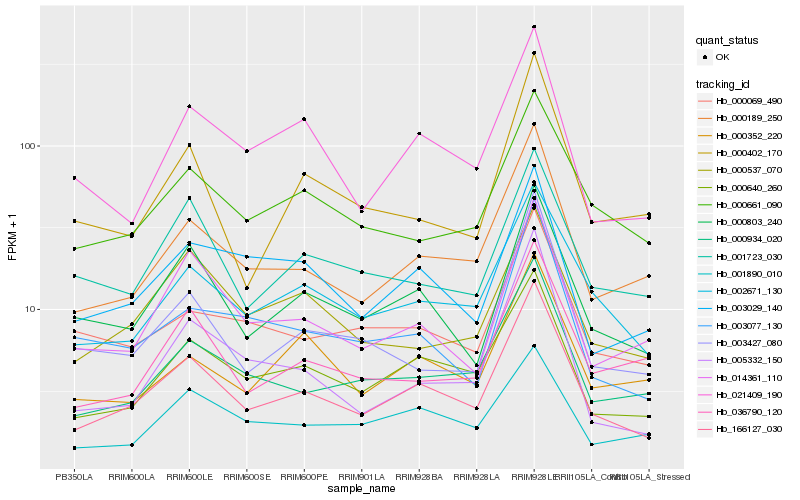
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_166127_030 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000640_260 |
0.089573767 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000189_250 |
0.0928721696 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002671_130 |
0.101503791 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 5 |
Hb_000803_240 |
0.1029497805 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 6 |
Hb_000934_020 |
0.1058496318 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 7 |
Hb_021409_190 |
0.1178702597 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 8 |
Hb_014361_110 |
0.1199824484 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_003029_140 |
0.1256840567 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000661_090 |
0.1257941658 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003077_130 |
0.1270069113 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 12 |
Hb_003427_080 |
0.1285778335 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_036790_120 |
0.1296659867 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005332_150 |
0.1301327592 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 15 |
Hb_000537_070 |
0.1311345528 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 16 |
Hb_000069_490 |
0.1316272821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001723_030 |
0.1327703077 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000352_220 |
0.1329083566 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001890_010 |
0.1338978362 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 20 |
Hb_000402_170 |
0.1348294922 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |