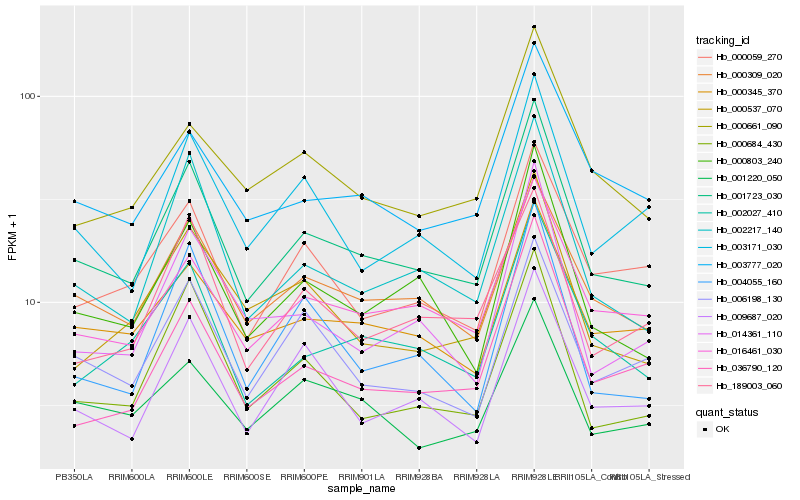
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001723_030 |
0.0 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002217_140 |
0.0796772394 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 3 |
Hb_000803_240 |
0.0810148044 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 4 |
Hb_000309_020 |
0.0850451481 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 5 |
Hb_016461_030 |
0.0885514149 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_002027_410 |
0.089275915 |
- |
- |
PREDICTED: protease Do-like 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000684_430 |
0.0947275504 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000345_370 |
0.0957082998 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 9 |
Hb_003777_020 |
0.0983462111 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 10 |
Hb_003171_030 |
0.0998694931 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 11 |
Hb_009687_020 |
0.0999691957 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 12 |
Hb_189003_060 |
0.1005542132 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000661_090 |
0.1010251674 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 14 |
Hb_014361_110 |
0.1015761371 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001220_050 |
0.1016567529 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 16 |
Hb_000059_270 |
0.1025563829 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 17 |
Hb_000537_070 |
0.1026618974 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 18 |
Hb_006198_130 |
0.1032413122 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 19 |
Hb_036790_120 |
0.105491075 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004055_160 |
0.1055279067 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |