| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
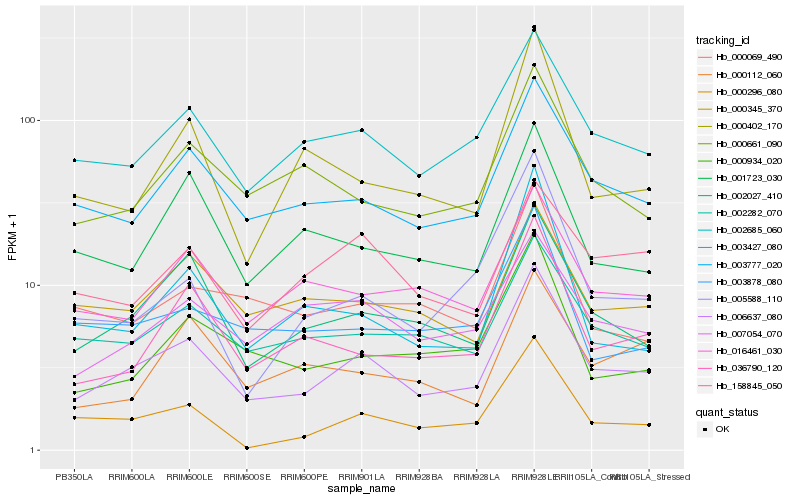
Hb_006637_080 |
0.0 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003777_020 |
0.107604947 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 3 |
Hb_002027_410 |
0.1078347692 |
- |
- |
PREDICTED: protease Do-like 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002685_060 |
0.1100381355 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 5 |
Hb_002282_070 |
0.1277658787 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 6 |
Hb_036790_120 |
0.1284568942 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 7 |
Hb_016461_030 |
0.1362041392 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_000661_090 |
0.136761277 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000296_080 |
0.1368862624 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35130 [Jatropha curcas] |
| 10 |
Hb_000934_020 |
0.1403135119 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 11 |
Hb_000345_370 |
0.1405510886 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 12 |
Hb_007054_070 |
0.1421148663 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 13 |
Hb_001723_030 |
0.1454852662 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003878_080 |
0.1479162883 |
- |
- |
PREDICTED: uncharacterized protein LOC105637045 [Jatropha curcas] |
| 15 |
Hb_158845_050 |
0.1503788995 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 16 |
Hb_000069_490 |
0.1527205219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000402_170 |
0.1539816962 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 18 |
Hb_000112_060 |
0.1552434097 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003427_080 |
0.1553433158 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005588_110 |
0.1555180578 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |