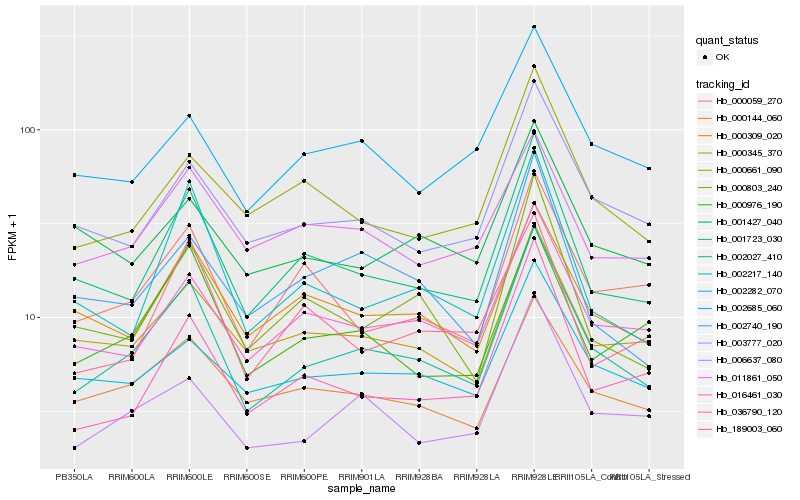
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_410 |
0.0 |
- |
- |
PREDICTED: protease Do-like 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001723_030 |
0.089275915 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_016461_030 |
0.1067913912 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_006637_080 |
0.1078347692 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003777_020 |
0.109329768 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 6 |
Hb_000803_240 |
0.1132915871 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 7 |
Hb_000144_060 |
0.1150303184 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002282_070 |
0.1162185712 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 9 |
Hb_000345_370 |
0.1169489402 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 10 |
Hb_002217_140 |
0.1178627294 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 11 |
Hb_000309_020 |
0.1186730979 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 12 |
Hb_000059_270 |
0.121380281 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 13 |
Hb_036790_120 |
0.1223406604 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000976_190 |
0.1227550851 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 15 |
Hb_000661_090 |
0.1230248742 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002685_060 |
0.1244579465 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 17 |
Hb_002740_190 |
0.1282779986 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_011861_050 |
0.1299994633 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_189003_060 |
0.1328459722 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001427_040 |
0.134927054 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |