| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
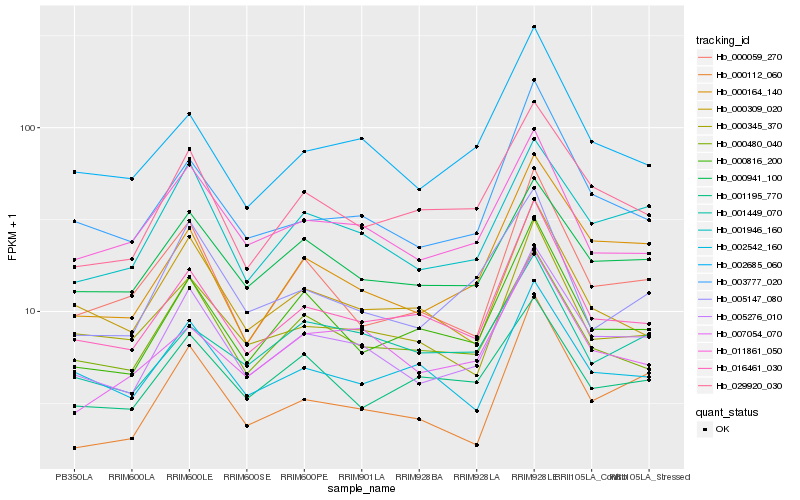
Hb_005276_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 2 |
Hb_001946_160 |
0.0634712464 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 3 |
Hb_000941_100 |
0.0824924225 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 4 |
Hb_000164_140 |
0.0867504567 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_029920_030 |
0.0868841388 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_016461_030 |
0.0888469153 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_000816_200 |
0.0917738593 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 8 |
Hb_003777_020 |
0.0922023502 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 9 |
Hb_000480_040 |
0.0940084833 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 10 |
Hb_011861_050 |
0.0974721963 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_002685_060 |
0.1013391446 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 12 |
Hb_000345_370 |
0.101457347 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 13 |
Hb_005147_080 |
0.1040540476 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000059_270 |
0.1045469461 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 15 |
Hb_000309_020 |
0.1048099801 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 16 |
Hb_000112_060 |
0.1057305287 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002542_160 |
0.1060451828 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001195_770 |
0.1094274624 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 19 |
Hb_007054_070 |
0.1115050966 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 20 |
Hb_001449_070 |
0.1133605081 |
- |
- |
conserved hypothetical protein [Ricinus communis] |