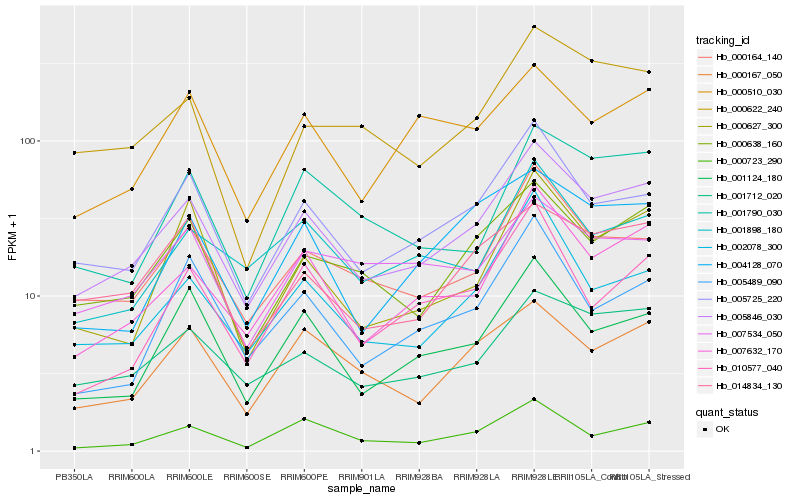
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005846_030 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007632_170 |
0.0849600033 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 3 |
Hb_005725_220 |
0.0879343004 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 4 |
Hb_001712_020 |
0.0944187945 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 5 |
Hb_000164_140 |
0.0955897538 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 6 |
Hb_000638_160 |
0.0956103475 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 7 |
Hb_000167_050 |
0.111748696 |
- |
- |
PREDICTED: uncharacterized protein LOC105641106 [Jatropha curcas] |
| 8 |
Hb_001124_180 |
0.1125562754 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 9 |
Hb_002078_300 |
0.1126574975 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 10 |
Hb_014834_130 |
0.113454121 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_004128_070 |
0.1191309398 |
- |
- |
PREDICTED: thioredoxin-like protein AAED1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001898_180 |
0.1203006741 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000622_240 |
0.1211898223 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000510_030 |
0.1215658339 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 15 |
Hb_000627_300 |
0.1233270265 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 16 |
Hb_000723_290 |
0.1243222536 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 17 |
Hb_001790_030 |
0.1261533331 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 18 |
Hb_010577_040 |
0.1274476597 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |
| 19 |
Hb_005489_090 |
0.1282837413 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 20 |
Hb_007534_050 |
0.1297268539 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |