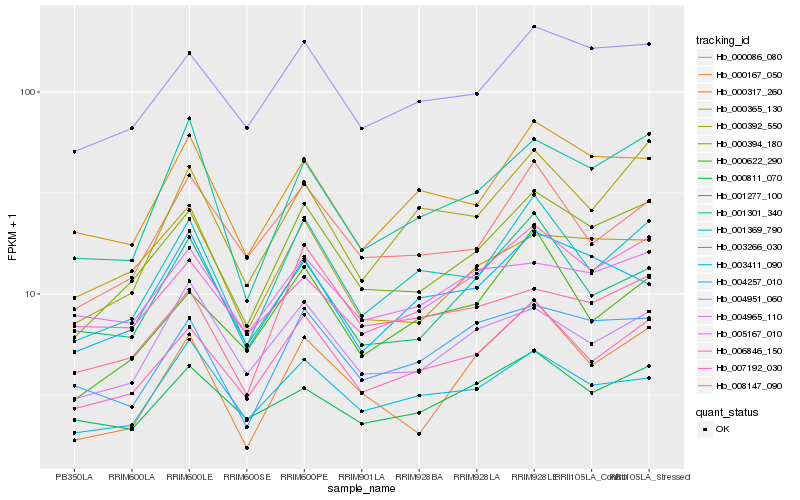
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000394_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 2 |
Hb_004951_060 |
0.0685599512 |
- |
- |
PREDICTED: uncharacterized protein LOC105634399 [Jatropha curcas] |
| 3 |
Hb_007192_030 |
0.0697928612 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 4 |
Hb_001369_790 |
0.0817787806 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001301_340 |
0.0951157629 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 6 |
Hb_006846_150 |
0.0951224887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004257_010 |
0.0984825394 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 8 |
Hb_004965_110 |
0.0999761445 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 9 |
Hb_000317_260 |
0.1007393301 |
- |
- |
unknown [Populus trichocarpa] |
| 10 |
Hb_000086_080 |
0.1013587368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000167_050 |
0.1047880315 |
- |
- |
PREDICTED: uncharacterized protein LOC105641106 [Jatropha curcas] |
| 12 |
Hb_003266_030 |
0.1053845719 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 13 |
Hb_000365_130 |
0.1080250806 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 14 |
Hb_003411_090 |
0.1081109301 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_001277_100 |
0.1084495176 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 16 |
Hb_000392_550 |
0.1094981198 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_005167_010 |
0.1098694264 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000811_070 |
0.1107078554 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 19 |
Hb_008147_090 |
0.1123226591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000622_290 |
0.1123336219 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |