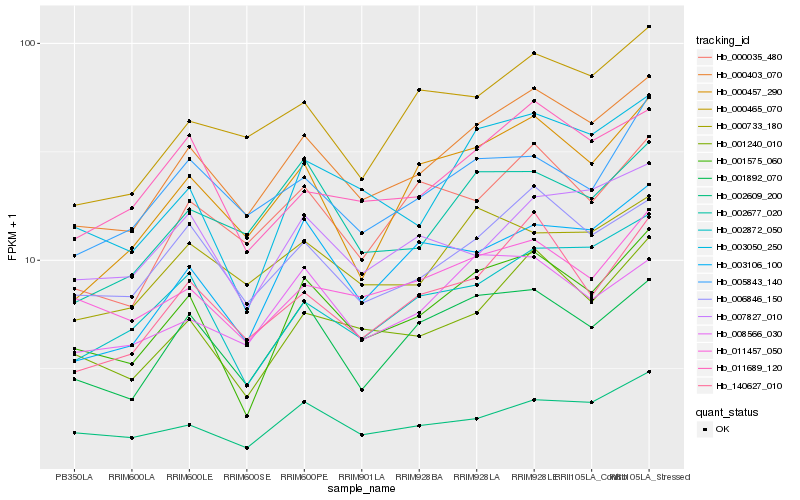
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000403_070 |
0.0 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_006846_150 |
0.0736684489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_140627_010 |
0.0808641831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001575_060 |
0.0826091552 |
- |
- |
PREDICTED: 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_001240_010 |
0.0826765639 |
- |
- |
PREDICTED: uncharacterized protein LOC105634101 [Jatropha curcas] |
| 6 |
Hb_000465_070 |
0.0835251614 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase small subunit 3-like [Jatropha curcas] |
| 7 |
Hb_000457_290 |
0.0856609965 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 8 |
Hb_001892_070 |
0.0887232016 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 9 |
Hb_011689_120 |
0.0889507286 |
- |
- |
PREDICTED: uncharacterized protein At4g15545 [Jatropha curcas] |
| 10 |
Hb_000035_480 |
0.0894056369 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 11 |
Hb_002609_200 |
0.0921883165 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_003050_250 |
0.0926517813 |
- |
- |
PREDICTED: uncharacterized protein At2g34460, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000733_180 |
0.0939729433 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 14 |
Hb_002872_050 |
0.0941469968 |
- |
- |
PREDICTED: thioredoxin-like 3-2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002677_020 |
0.0952285938 |
- |
- |
PREDICTED: uncharacterized protein LOC105642265 [Jatropha curcas] |
| 16 |
Hb_008566_030 |
0.0999466105 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 17 |
Hb_003106_100 |
0.1011767338 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 18 |
Hb_011457_050 |
0.1018666725 |
- |
- |
PREDICTED: TP53-regulating kinase [Jatropha curcas] |
| 19 |
Hb_005843_140 |
0.1020574045 |
- |
- |
Red chlorophyll catabolite reductase, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_007827_010 |
0.1040643276 |
- |
- |
PREDICTED: uncharacterized protein C12B10.15c [Jatropha curcas] |