| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
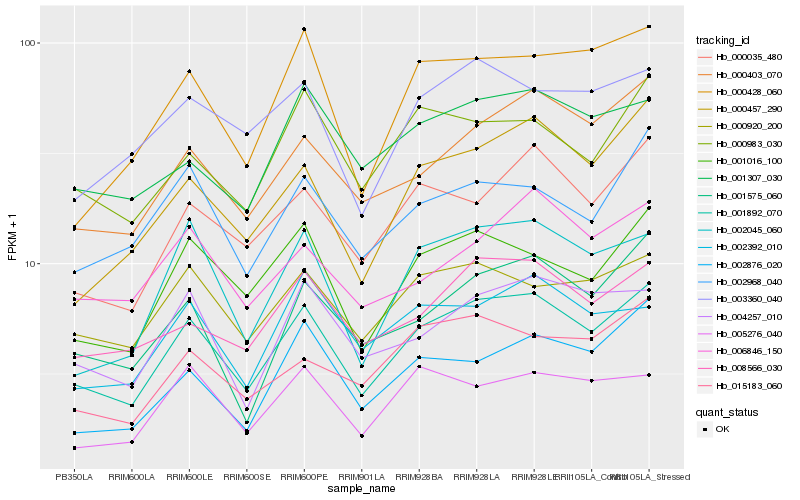
Hb_001892_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 2 |
Hb_001016_100 |
0.0824273848 |
- |
- |
PREDICTED: V-type proton ATPase subunit E-like [Pyrus x bretschneideri] |
| 3 |
Hb_000403_070 |
0.0887232016 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_002045_060 |
0.0895843569 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 5 |
Hb_008566_030 |
0.0899869761 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 6 |
Hb_000035_480 |
0.0902518195 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 7 |
Hb_000457_290 |
0.0909681091 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 8 |
Hb_000920_200 |
0.0913829461 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001575_060 |
0.0956421068 |
- |
- |
PREDICTED: 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_004257_010 |
0.0963249259 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000428_060 |
0.098580742 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_002968_040 |
0.0989332184 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7-like [Jatropha curcas] |
| 13 |
Hb_015183_060 |
0.1002682275 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 14 |
Hb_000983_030 |
0.1008316724 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 15 |
Hb_001307_030 |
0.1016291493 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003360_040 |
0.1030197414 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 17 |
Hb_006846_150 |
0.104063587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002876_020 |
0.104529797 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002392_010 |
0.1061426238 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 20 |
Hb_005276_040 |
0.1066635139 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |