| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
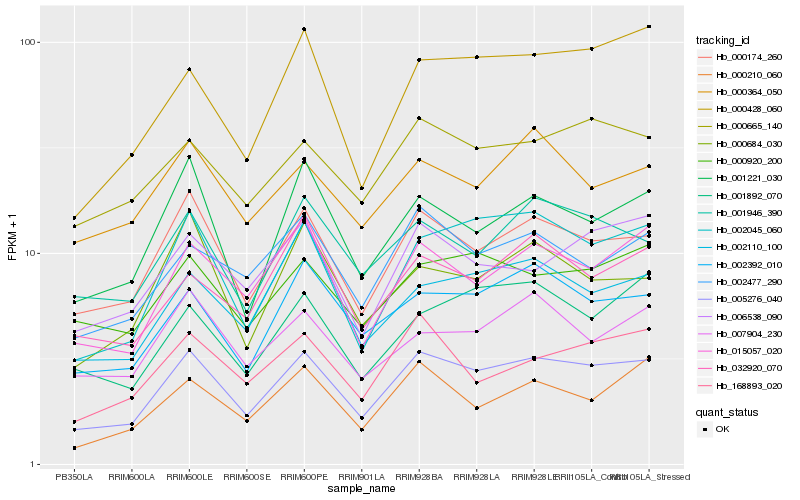
Hb_005276_040 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 2 |
Hb_000174_260 |
0.0650859371 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 3 |
Hb_002045_060 |
0.0746980964 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 4 |
Hb_015057_020 |
0.0872707652 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 5 |
Hb_002392_010 |
0.0876001064 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 6 |
Hb_000428_060 |
0.0899764079 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_001221_030 |
0.0945761906 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 8 |
Hb_002477_290 |
0.0955536787 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_001946_390 |
0.0968734359 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 10 |
Hb_000665_140 |
0.0969183603 |
- |
- |
PREDICTED: protein MEMO1 [Jatropha curcas] |
| 11 |
Hb_006538_090 |
0.0990221799 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 12 |
Hb_000920_200 |
0.0999703641 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_168893_020 |
0.1023408857 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 14 |
Hb_000210_060 |
0.1042793789 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 15 |
Hb_032920_070 |
0.1066292947 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 16 |
Hb_007904_230 |
0.106648234 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001892_070 |
0.1066635139 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 18 |
Hb_000684_030 |
0.1077503176 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 19 |
Hb_000364_050 |
0.1078476097 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002110_100 |
0.1086707676 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |