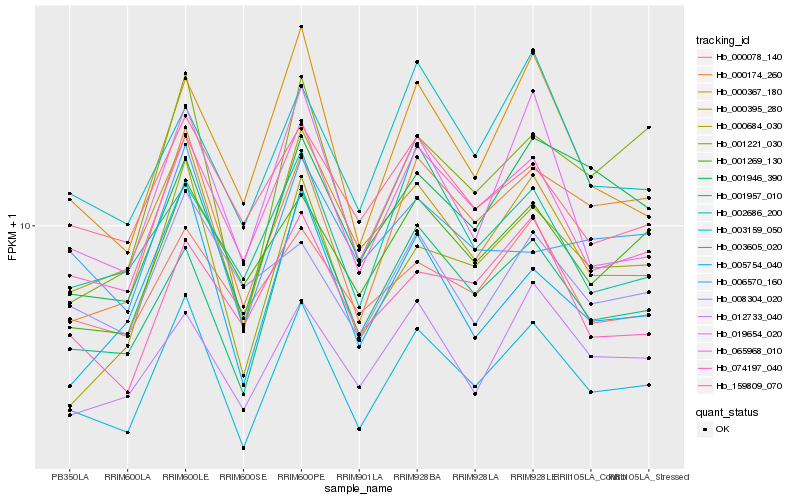
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003605_020 |
0.0 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 2 |
Hb_065968_010 |
0.0641312661 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000395_280 |
0.0694890668 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 4 |
Hb_001957_010 |
0.0728934949 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000174_260 |
0.0748074179 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 6 |
Hb_001269_130 |
0.0828582319 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 7 |
Hb_019654_020 |
0.0858027137 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 8 |
Hb_012733_040 |
0.0872052005 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 9 |
Hb_005754_040 |
0.0880245377 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000367_180 |
0.0888351714 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 11 |
Hb_159809_070 |
0.0894337829 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 12 |
Hb_000684_030 |
0.0902370991 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 13 |
Hb_000078_140 |
0.0925964231 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 14 |
Hb_001946_390 |
0.0928052429 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 15 |
Hb_003159_050 |
0.0934655332 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_074197_040 |
0.0942034348 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 17 |
Hb_002686_200 |
0.0961100839 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 18 |
Hb_006570_160 |
0.0965112624 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 19 |
Hb_001221_030 |
0.0973809657 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 20 |
Hb_008304_020 |
0.0991055744 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |