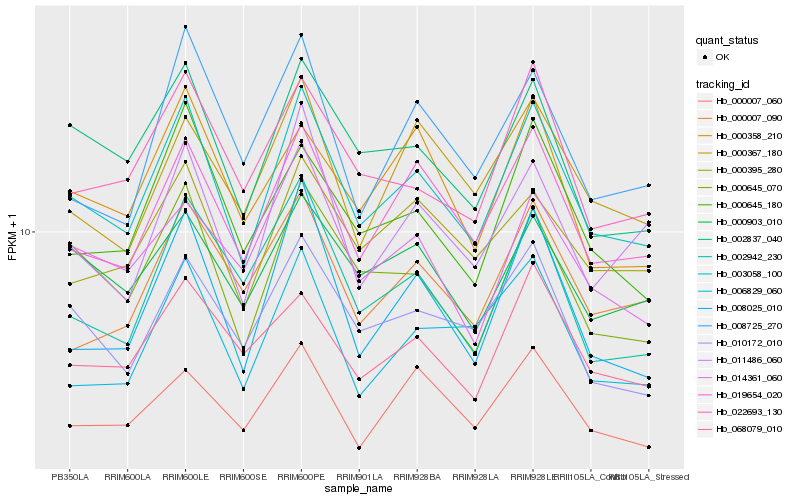
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003058_100 |
0.0 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_019654_020 |
0.0745902573 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 3 |
Hb_002837_040 |
0.0829163096 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 4 |
Hb_000645_070 |
0.0833220333 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_022693_130 |
0.0906616284 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000645_180 |
0.0913477878 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 7 |
Hb_010172_010 |
0.0916790159 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 8 |
Hb_008025_010 |
0.0933671832 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |
| 9 |
Hb_011486_060 |
0.0944961595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_068079_010 |
0.0973258703 |
- |
- |
- |
| 11 |
Hb_000395_280 |
0.0977592931 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 12 |
Hb_000007_090 |
0.0998710316 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 13 |
Hb_000903_010 |
0.1001555849 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 14 |
Hb_002942_230 |
0.1008187543 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 15 |
Hb_008725_270 |
0.100891437 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 16 |
Hb_006829_060 |
0.1015543199 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000358_210 |
0.1028014002 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 18 |
Hb_014361_060 |
0.1034528551 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 19 |
Hb_000007_060 |
0.1038438707 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000367_180 |
0.104143338 |
- |
- |
Heparanase-2, putative [Ricinus communis] |