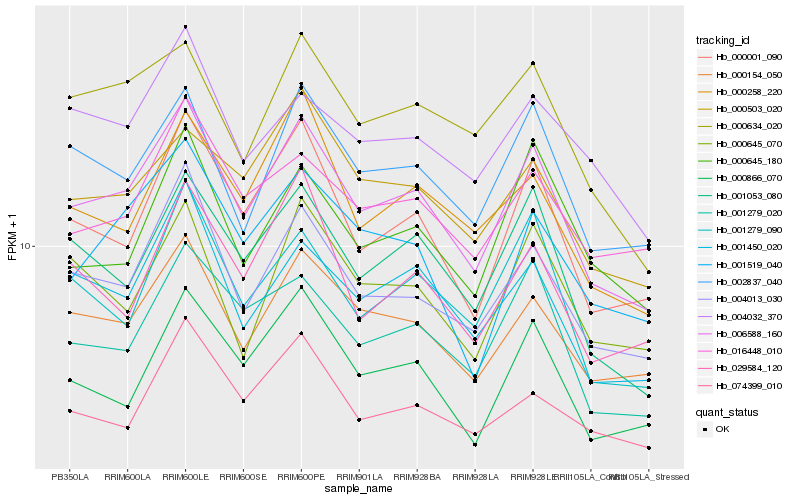
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006588_160 |
0.0 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_000154_050 |
0.0721608378 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 3 |
Hb_004013_030 |
0.0732176868 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 4 |
Hb_000001_090 |
0.074002964 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 5 |
Hb_001053_080 |
0.0844017698 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 6 |
Hb_002837_040 |
0.0846131421 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 7 |
Hb_001450_020 |
0.0881853433 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_001279_020 |
0.0900328613 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 9 |
Hb_000634_020 |
0.096351308 |
- |
- |
hypothetical protein JCGZ_11888 [Jatropha curcas] |
| 10 |
Hb_001519_040 |
0.0972740274 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 11 |
Hb_001279_090 |
0.0978098749 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 12 |
Hb_000258_220 |
0.0987438667 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000866_070 |
0.0998168968 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 14 |
Hb_000503_020 |
0.1012015911 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 15 |
Hb_016448_010 |
0.1014907488 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 16 |
Hb_074399_010 |
0.1047284127 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 17 |
Hb_000645_070 |
0.1051076041 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_004032_370 |
0.1063761513 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 19 |
Hb_000645_180 |
0.1067644044 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 20 |
Hb_029584_120 |
0.1071568003 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |