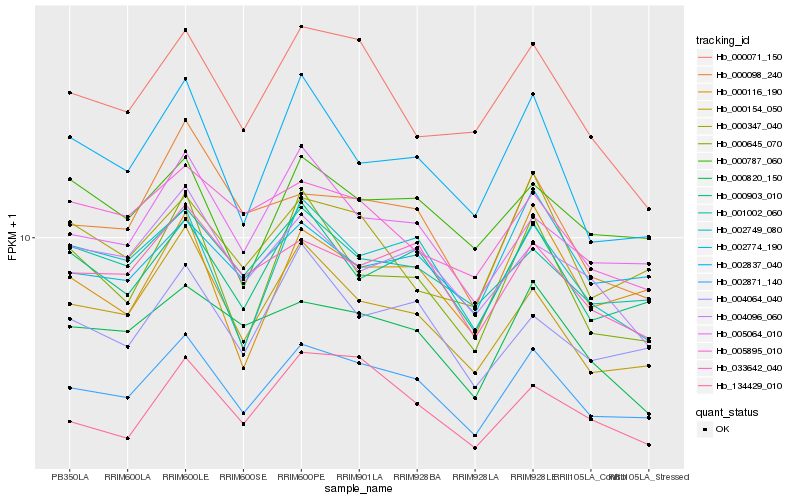
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002871_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 2 |
Hb_000903_010 |
0.0808988111 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 3 |
Hb_000154_050 |
0.0817214628 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 4 |
Hb_002749_080 |
0.0862947037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005064_010 |
0.0872231811 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 6 |
Hb_002837_040 |
0.0883382717 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 7 |
Hb_033642_040 |
0.0889470701 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000116_190 |
0.0948151843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000645_070 |
0.0971107076 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_004064_040 |
0.0975809742 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 11 |
Hb_000098_240 |
0.0983078131 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 12 |
Hb_001002_060 |
0.0990765758 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 13 |
Hb_000820_150 |
0.0995403799 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 14 |
Hb_000071_150 |
0.0998935907 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 15 |
Hb_000347_040 |
0.1017421229 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 16 |
Hb_000787_060 |
0.104122834 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_002774_190 |
0.1080231899 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 18 |
Hb_004096_060 |
0.1081503866 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 19 |
Hb_134429_010 |
0.1098578757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005895_010 |
0.1101883465 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |