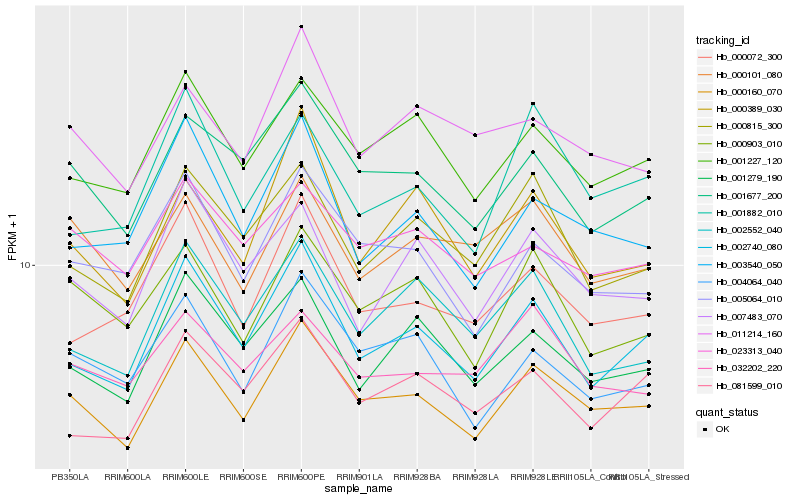
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000160_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 2 |
Hb_002740_080 |
0.073396332 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 3 |
Hb_001677_200 |
0.0754129191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001227_120 |
0.0792602076 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 5 |
Hb_004064_040 |
0.081585972 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 6 |
Hb_005064_010 |
0.0818087935 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 7 |
Hb_000903_010 |
0.0842918736 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 8 |
Hb_000815_300 |
0.0919221975 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 9 |
Hb_011214_160 |
0.0925670979 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 10 |
Hb_001279_190 |
0.0928353095 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003540_050 |
0.0937164697 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 12 |
Hb_000389_030 |
0.0942751668 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 13 |
Hb_000072_300 |
0.0970300791 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 14 |
Hb_023313_040 |
0.0977673186 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007483_070 |
0.0984867037 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 16 |
Hb_000101_080 |
0.0985043769 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 17 |
Hb_081599_010 |
0.1021207887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_032202_220 |
0.1022732989 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_002552_040 |
0.1025183656 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 20 |
Hb_001882_010 |
0.1035824048 |
- |
- |
- |