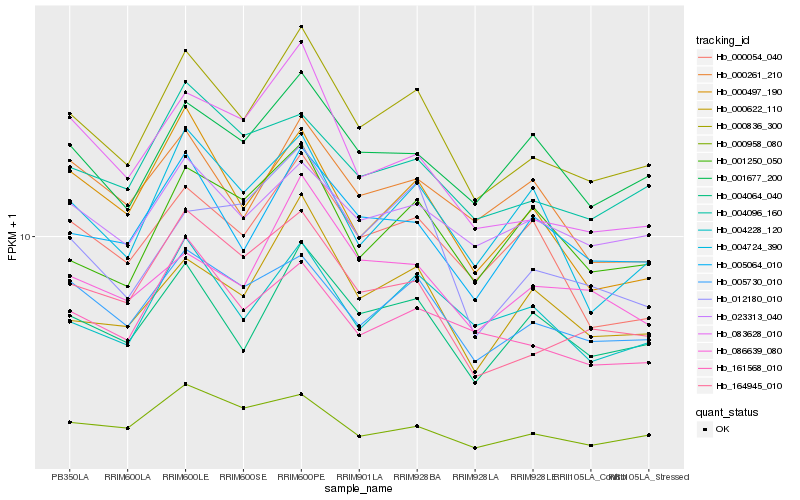
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_300 |
0.0 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 2 |
Hb_004064_040 |
0.0766553374 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 3 |
Hb_005730_010 |
0.084136048 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 4 |
Hb_164945_010 |
0.0884709477 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 5 |
Hb_005064_010 |
0.0893788428 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 6 |
Hb_000622_110 |
0.0900570719 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 7 |
Hb_004228_120 |
0.0917379107 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 8 |
Hb_001677_200 |
0.0955171153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_083628_010 |
0.0967487567 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 10 |
Hb_000054_040 |
0.0974653995 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 11 |
Hb_161568_010 |
0.0985073381 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 12 |
Hb_004724_390 |
0.1013326908 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 13 |
Hb_004096_160 |
0.1015439148 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 14 |
Hb_000497_190 |
0.1017641752 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 15 |
Hb_023313_040 |
0.1020044415 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001250_050 |
0.1027633512 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000958_080 |
0.1031753855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_012180_010 |
0.1031929523 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |
| 19 |
Hb_086639_080 |
0.1041668681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000261_210 |
0.1058403348 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |