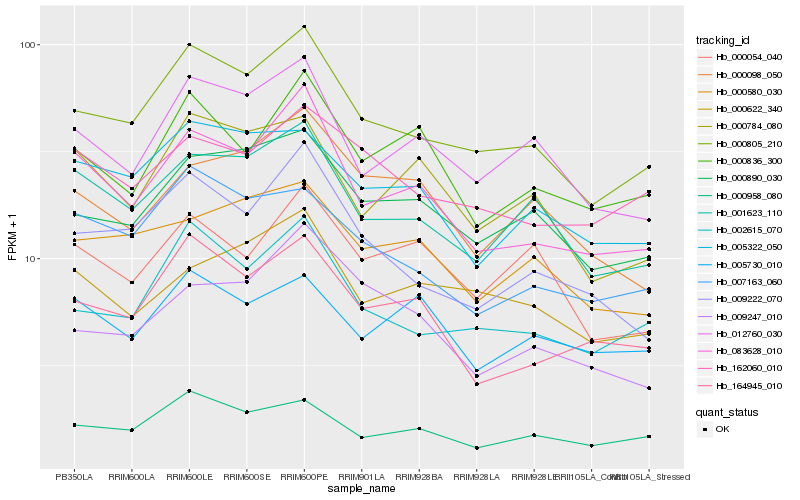
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083628_010 |
0.0 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 2 |
Hb_000805_210 |
0.0885118132 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 3 |
Hb_164945_010 |
0.095562028 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 4 |
Hb_001623_110 |
0.0961632785 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_000836_300 |
0.0967487567 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 6 |
Hb_012760_030 |
0.1007242924 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 7 |
Hb_009222_070 |
0.1058565334 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_000784_080 |
0.112496734 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 9 |
Hb_000622_340 |
0.1154671388 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 10 |
Hb_000098_050 |
0.1184282957 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 11 |
Hb_002615_070 |
0.1195813789 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000890_030 |
0.1205857092 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 13 |
Hb_005322_050 |
0.1210316504 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 14 |
Hb_009247_010 |
0.1222011564 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000958_080 |
0.1238550811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_162060_010 |
0.1243688962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007163_060 |
0.1249336841 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 18 |
Hb_000580_030 |
0.1257552449 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 19 |
Hb_000054_040 |
0.1273671236 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 20 |
Hb_005730_010 |
0.1274623867 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |