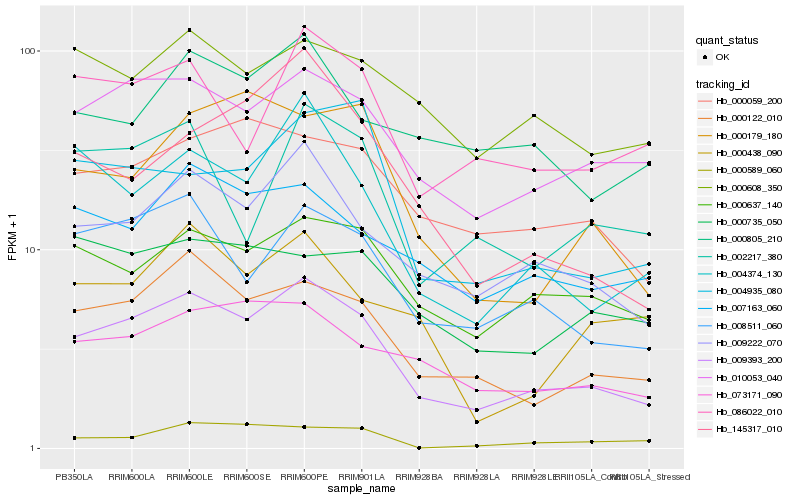
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009393_200 |
0.0 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 2 |
Hb_000122_010 |
0.1045901043 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_009222_070 |
0.1049720898 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_010053_040 |
0.1167527759 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 5 |
Hb_008511_060 |
0.1195636579 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 6 |
Hb_004374_130 |
0.1348576521 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 7 |
Hb_000637_140 |
0.1382489038 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 8 |
Hb_073171_090 |
0.1386438246 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 9 |
Hb_000059_200 |
0.1426721606 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 10 |
Hb_007163_060 |
0.143148746 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 11 |
Hb_086022_010 |
0.1514369583 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 12 |
Hb_004935_080 |
0.1541722928 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 13 |
Hb_002217_380 |
0.1546986254 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 14 |
Hb_000608_350 |
0.1555524122 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 15 |
Hb_000179_180 |
0.1562532017 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 16 |
Hb_000805_210 |
0.1597770472 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 17 |
Hb_000735_050 |
0.1600728266 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 18 |
Hb_145317_010 |
0.1603700558 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 19 |
Hb_000438_090 |
0.160813734 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 20 |
Hb_000589_060 |
0.1627135285 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00699 [Jatropha curcas] |