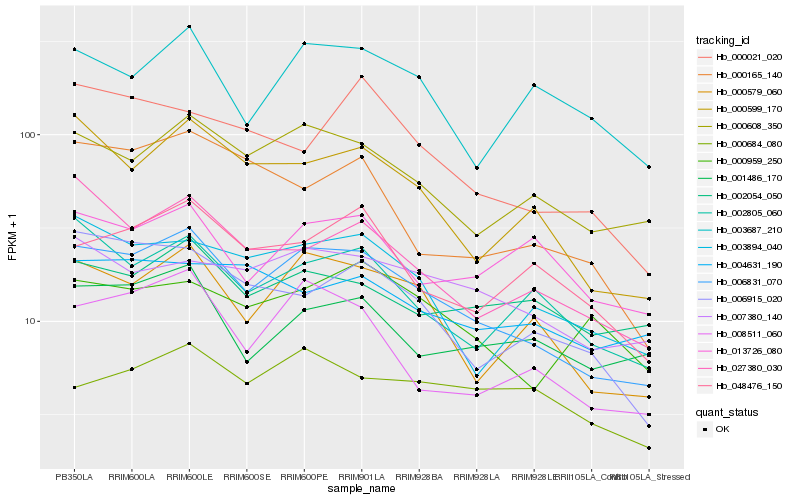
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006831_070 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase 4 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000579_060 |
0.0936661371 |
- |
- |
PREDICTED: uncharacterized protein LOC105633849 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000608_350 |
0.1048435412 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 4 |
Hb_000599_170 |
0.1062383495 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 5 |
Hb_008511_060 |
0.1097187451 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 6 |
Hb_003894_040 |
0.1177050018 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 7 |
Hb_007380_140 |
0.1190220328 |
- |
- |
PREDICTED: probable galacturonosyltransferase 4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_027380_030 |
0.1201534973 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002805_060 |
0.1203157046 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 10 |
Hb_006915_020 |
0.1259768424 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 11 |
Hb_048476_150 |
0.1273666142 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000165_140 |
0.1329917393 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 13 |
Hb_003687_210 |
0.13512377 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 14 |
Hb_000021_020 |
0.1352399473 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |
| 15 |
Hb_001486_170 |
0.13568298 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 16 |
Hb_013726_080 |
0.1390909557 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 17 |
Hb_002054_050 |
0.1396616832 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 18 |
Hb_000684_080 |
0.1406430357 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 19 |
Hb_004631_190 |
0.1425556337 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 20 |
Hb_000959_250 |
0.1430992799 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |