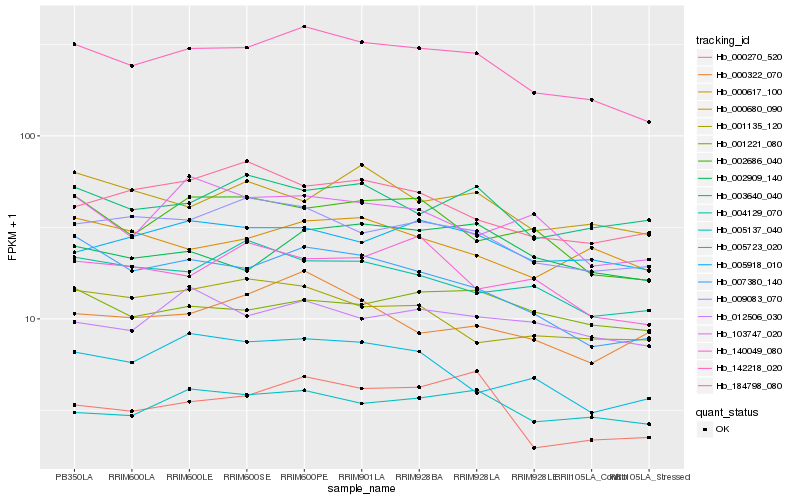
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142218_020 |
0.0 |
- |
- |
actin family protein [Populus trichocarpa] |
| 2 |
Hb_009083_070 |
0.0733237052 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 3 |
Hb_007380_140 |
0.0777379977 |
- |
- |
PREDICTED: probable galacturonosyltransferase 4 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002686_040 |
0.0820997944 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 5 |
Hb_005137_040 |
0.0834420414 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 6 |
Hb_005918_010 |
0.0875475236 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 7 |
Hb_004129_070 |
0.0875882547 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 8 |
Hb_000617_100 |
0.0881952961 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 9 |
Hb_003640_040 |
0.0886367915 |
- |
- |
PREDICTED: uncharacterized protein LOC105647348 [Jatropha curcas] |
| 10 |
Hb_005723_020 |
0.0909077634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001221_080 |
0.092503603 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_012506_030 |
0.0927179033 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 13 |
Hb_000322_070 |
0.0930018603 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_000270_520 |
0.0932213597 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_140049_080 |
0.0936686756 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 16 |
Hb_000680_090 |
0.0946552462 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 17 |
Hb_001135_120 |
0.0946605482 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002909_140 |
0.0951054392 |
- |
- |
PREDICTED: D-xylose-proton symporter-like 2 [Jatropha curcas] |
| 19 |
Hb_184798_080 |
0.0957770558 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 20 |
Hb_103747_020 |
0.0958184543 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |