| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
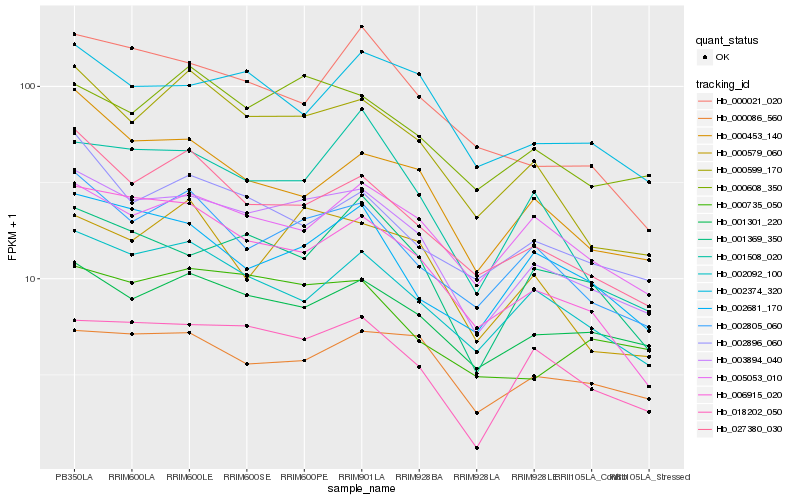
Hb_003894_040 |
0.0 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 2 |
Hb_002805_060 |
0.0876926306 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 3 |
Hb_000599_170 |
0.0929158391 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 4 |
Hb_018202_050 |
0.0945207066 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A [Jatropha curcas] |
| 5 |
Hb_027380_030 |
0.0957540409 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_001301_220 |
0.096212507 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 7 |
Hb_006915_020 |
0.0970562836 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 8 |
Hb_002092_100 |
0.0999542525 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 9 |
Hb_000608_350 |
0.1006907165 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 10 |
Hb_000735_050 |
0.1050670625 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 11 |
Hb_001369_350 |
0.108213539 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 12 |
Hb_000086_560 |
0.1126838662 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002374_320 |
0.1130836297 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 14 |
Hb_002681_170 |
0.1132123833 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 15 |
Hb_001508_020 |
0.1145290754 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 16 |
Hb_002896_060 |
0.1148832249 |
- |
- |
PREDICTED: synaptotagmin-2 [Jatropha curcas] |
| 17 |
Hb_000453_140 |
0.1153448408 |
- |
- |
hypothetical protein POPTR_0009s02600g [Populus trichocarpa] |
| 18 |
Hb_000579_060 |
0.1153540181 |
- |
- |
PREDICTED: uncharacterized protein LOC105633849 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005053_010 |
0.1164691016 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 20 |
Hb_000021_020 |
0.1172162757 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |