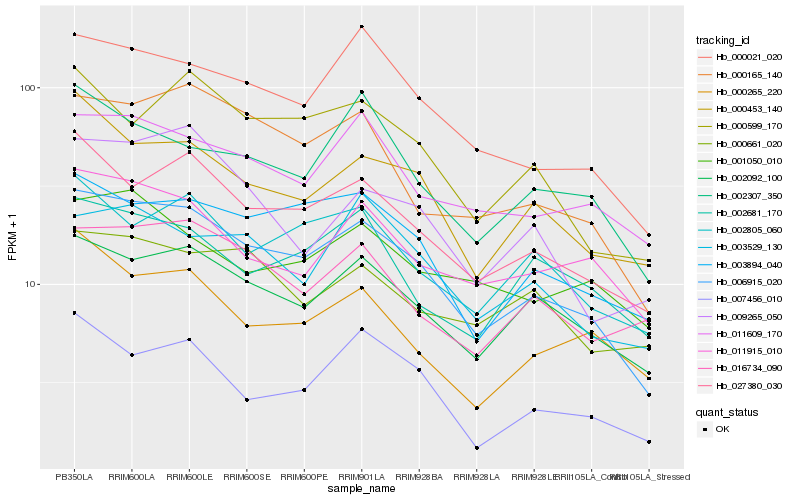
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006915_020 |
0.0 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 2 |
Hb_027380_030 |
0.0830330188 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_002092_100 |
0.088117211 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 4 |
Hb_000021_020 |
0.0907716433 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |
| 5 |
Hb_000165_140 |
0.0965194711 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 6 |
Hb_003894_040 |
0.0970562836 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 7 |
Hb_000453_140 |
0.1004171114 |
- |
- |
hypothetical protein POPTR_0009s02600g [Populus trichocarpa] |
| 8 |
Hb_000599_170 |
0.1052288409 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 9 |
Hb_011609_170 |
0.1096450931 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 10 |
Hb_002805_060 |
0.1110186158 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 11 |
Hb_011915_010 |
0.1110366124 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 12 |
Hb_007456_010 |
0.1126556622 |
- |
- |
hypothetical protein L484_003540 [Morus notabilis] |
| 13 |
Hb_002307_350 |
0.1134854912 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 14 |
Hb_003529_130 |
0.1163846324 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_009265_050 |
0.1183391779 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 16 |
Hb_016734_090 |
0.1193224918 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 17 |
Hb_002681_170 |
0.1215298756 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 18 |
Hb_000661_020 |
0.1232421664 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 19 |
Hb_000265_220 |
0.1236972771 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 20 |
Hb_001050_010 |
0.1248274049 |
- |
- |
Advillin [Gossypium arboreum] |