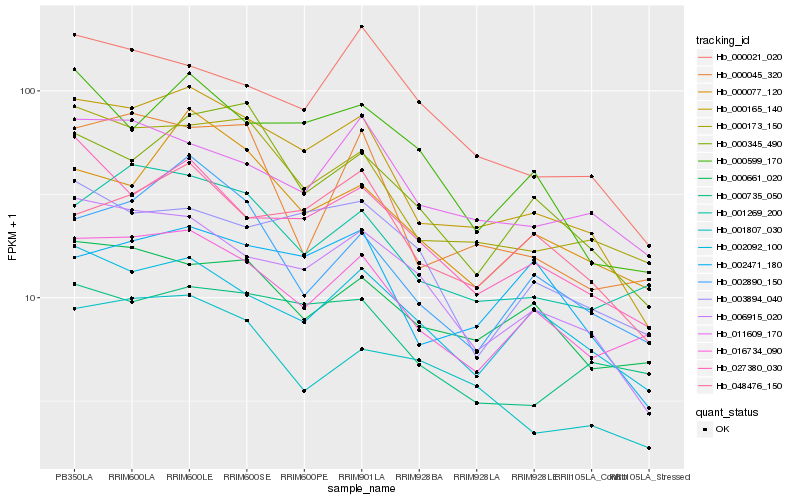
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000165_140 |
0.0 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 2 |
Hb_000173_150 |
0.0922378678 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 3 |
Hb_006915_020 |
0.0965194711 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 4 |
Hb_027380_030 |
0.1112441492 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_016734_090 |
0.1144404342 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 6 |
Hb_000599_170 |
0.1165648543 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 7 |
Hb_002092_100 |
0.1200691887 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 8 |
Hb_000345_490 |
0.1203362644 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 9 |
Hb_000077_120 |
0.1249296921 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 10 |
Hb_001269_200 |
0.1249787853 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 11 |
Hb_002471_180 |
0.1252879671 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_048476_150 |
0.1258576583 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000735_050 |
0.1281470445 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 14 |
Hb_000021_020 |
0.1288020167 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |
| 15 |
Hb_002890_150 |
0.1295260628 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000045_320 |
0.1295864979 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011609_170 |
0.1301497887 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 18 |
Hb_001807_030 |
0.1304592782 |
- |
- |
PREDICTED: CDT1-like protein b [Jatropha curcas] |
| 19 |
Hb_000661_020 |
0.1314154812 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 20 |
Hb_003894_040 |
0.131994695 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |