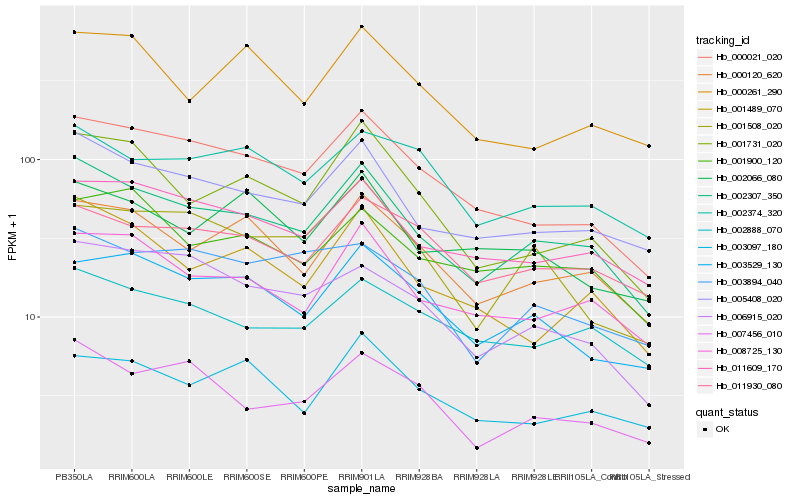
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000021_020 |
0.0 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |
| 2 |
Hb_003529_130 |
0.0811769057 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_011609_170 |
0.0820228153 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 4 |
Hb_002307_350 |
0.0898810048 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 5 |
Hb_006915_020 |
0.0907716433 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 6 |
Hb_005408_020 |
0.0947762448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008725_130 |
0.0998994735 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 8 |
Hb_001731_020 |
0.1008196829 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 9 |
Hb_000120_620 |
0.1010189755 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 10 |
Hb_001489_070 |
0.1061760946 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 11 |
Hb_003097_180 |
0.1079409418 |
- |
- |
PREDICTED: crossover junction endonuclease MUS81 [Jatropha curcas] |
| 12 |
Hb_011930_080 |
0.1109276615 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 13 |
Hb_002374_320 |
0.1111219776 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 14 |
Hb_001508_020 |
0.1113974556 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 15 |
Hb_002888_070 |
0.1146558817 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 16 |
Hb_000261_290 |
0.1163753833 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 17 |
Hb_003894_040 |
0.1172162757 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 18 |
Hb_001900_120 |
0.1177877633 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 19 |
Hb_002066_080 |
0.1180951284 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 20 |
Hb_007456_010 |
0.1186969321 |
- |
- |
hypothetical protein L484_003540 [Morus notabilis] |