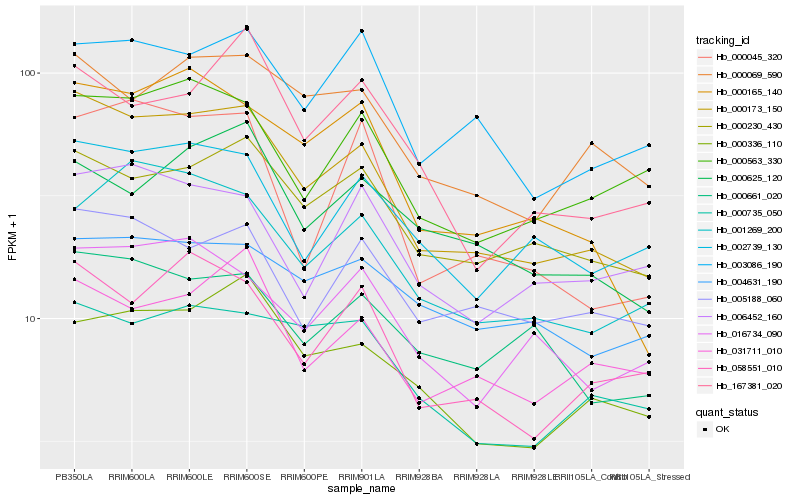
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000173_150 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 2 |
Hb_000165_140 |
0.0922378678 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 3 |
Hb_058551_010 |
0.092359123 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_000230_430 |
0.0953303606 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_000069_590 |
0.0981087453 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 6 |
Hb_002739_130 |
0.1013413336 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 7 |
Hb_000735_050 |
0.1020770673 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 8 |
Hb_001269_200 |
0.1021748303 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 9 |
Hb_016734_090 |
0.1022974366 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 10 |
Hb_000336_110 |
0.1026448382 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000661_020 |
0.1030890003 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 12 |
Hb_003086_190 |
0.105129682 |
- |
- |
PREDICTED: uncharacterized protein LOC105635794 [Jatropha curcas] |
| 13 |
Hb_031711_010 |
0.1070225289 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000045_320 |
0.1070503647 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005188_060 |
0.1078845388 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 16 |
Hb_000563_330 |
0.1079533697 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000625_120 |
0.1113623214 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 18 |
Hb_167381_020 |
0.1141814875 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 19 |
Hb_004631_190 |
0.1144570767 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 20 |
Hb_006452_160 |
0.1157604514 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |