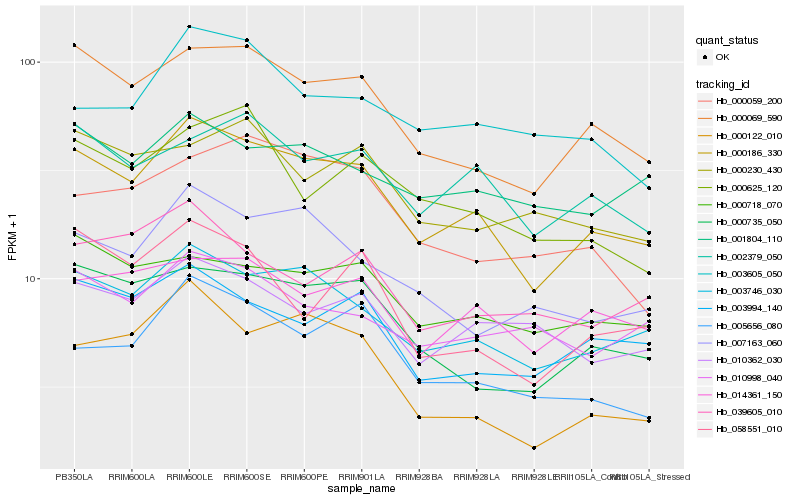
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_330 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 2 |
Hb_003746_030 |
0.0681415201 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 3 |
Hb_000069_590 |
0.0758142414 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 4 |
Hb_000735_050 |
0.0972972928 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 5 |
Hb_002379_050 |
0.098425223 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 6 |
Hb_014361_150 |
0.0999405232 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 7 |
Hb_003994_140 |
0.1040018163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007163_060 |
0.1041621534 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 9 |
Hb_005656_080 |
0.10422712 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 10 |
Hb_000718_070 |
0.1050985727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010362_030 |
0.1102281178 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_001804_110 |
0.1116041048 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 13 |
Hb_010998_040 |
0.1163686039 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 14 |
Hb_000625_120 |
0.11657773 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 15 |
Hb_003605_050 |
0.1171902567 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000059_200 |
0.1177514397 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 17 |
Hb_039605_010 |
0.1190369261 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000230_430 |
0.1191281373 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_058551_010 |
0.1192072219 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_000122_010 |
0.1194476918 |
- |
- |
amino acid transporter, putative [Ricinus communis] |