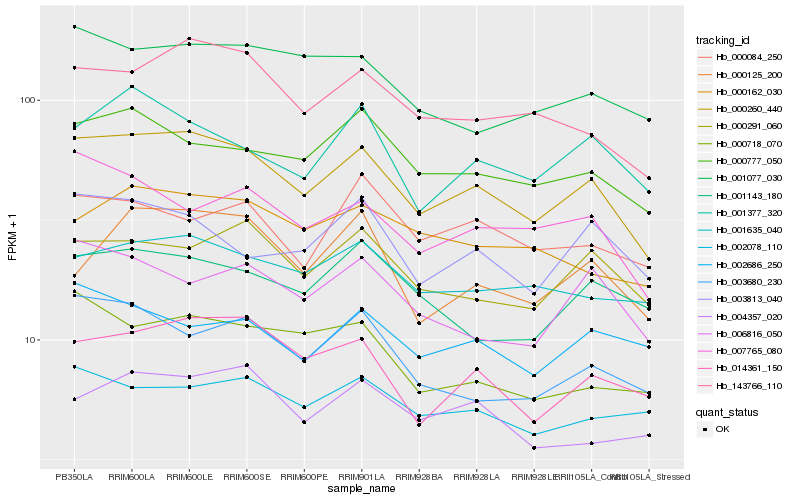
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_440 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 2 |
Hb_143766_110 |
0.0699742621 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 3 |
Hb_000777_050 |
0.0748887597 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 4 |
Hb_014361_150 |
0.0767780154 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 5 |
Hb_000291_060 |
0.0799270039 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 6 |
Hb_007765_080 |
0.0808311489 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001635_040 |
0.0817957876 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_001377_320 |
0.0818136243 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 9 |
Hb_003813_040 |
0.0843156776 |
- |
- |
PREDICTED: elongator complex protein 5 [Jatropha curcas] |
| 10 |
Hb_003680_230 |
0.0856642599 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 11 |
Hb_000162_030 |
0.0862272394 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_006816_050 |
0.0870879577 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 13 |
Hb_001077_030 |
0.0879937431 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004357_020 |
0.0888573042 |
- |
- |
PREDICTED: uncharacterized protein LOC105637355 [Jatropha curcas] |
| 15 |
Hb_000718_070 |
0.0901055363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000125_200 |
0.0910004176 |
transcription factor |
TF Family: bZIP |
PREDICTED: probable transcription factor PosF21 [Jatropha curcas] |
| 17 |
Hb_001143_180 |
0.0924648331 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 18 |
Hb_002686_250 |
0.0925008483 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_000084_250 |
0.093276776 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 20 |
Hb_002078_110 |
0.0932921086 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |