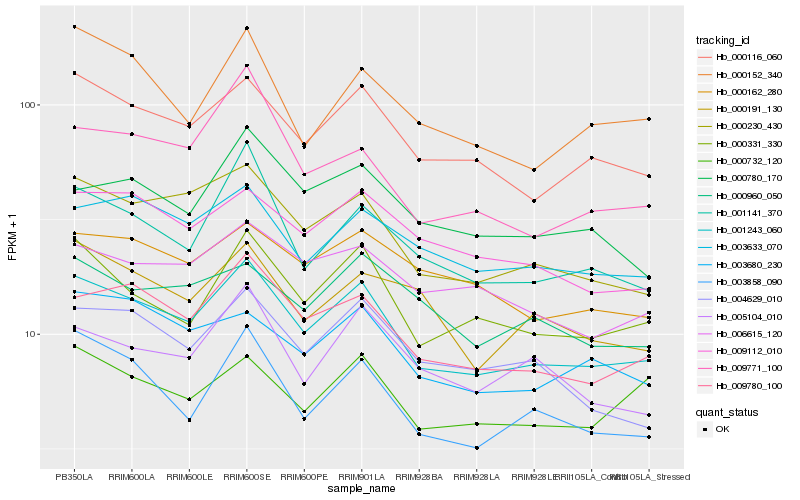
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001243_060 |
0.0 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 2 |
Hb_000116_060 |
0.0564725153 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 3 |
Hb_009780_100 |
0.0580869465 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 4 |
Hb_000230_430 |
0.0587289964 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_000331_330 |
0.0671802676 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003858_090 |
0.0731276208 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 7 |
Hb_001141_370 |
0.0740885493 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 8 |
Hb_003680_230 |
0.077998453 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 9 |
Hb_003633_070 |
0.0795066286 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 10 |
Hb_009112_010 |
0.0801974778 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 11 |
Hb_009771_100 |
0.083162254 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 12 |
Hb_004629_010 |
0.0848961152 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 13 |
Hb_006615_120 |
0.0850691074 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 14 |
Hb_000162_280 |
0.0851743618 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 15 |
Hb_000152_340 |
0.0860319864 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 16 |
Hb_005104_010 |
0.087939206 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 17 |
Hb_000960_050 |
0.0884136341 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 18 |
Hb_000780_170 |
0.0885470811 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 19 |
Hb_000732_120 |
0.0886513812 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000191_130 |
0.0890191206 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |