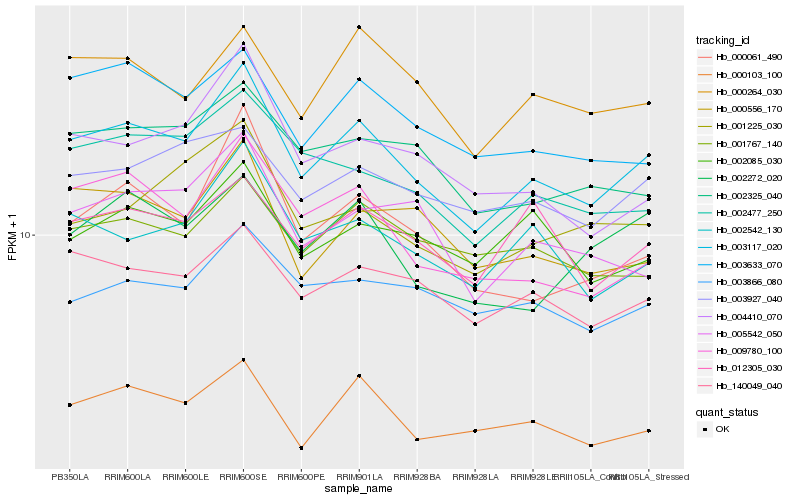
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003117_020 |
0.0 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 2 |
Hb_140049_040 |
0.0709792374 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 3 |
Hb_002542_130 |
0.0733219643 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 4 |
Hb_000103_100 |
0.0755495662 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_002477_250 |
0.0770175606 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 6 |
Hb_009780_100 |
0.0774499672 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 7 |
Hb_003633_070 |
0.0791492695 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 8 |
Hb_000264_030 |
0.0795244592 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 9 |
Hb_002325_040 |
0.0800493869 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003927_040 |
0.0809132094 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 11 |
Hb_000556_170 |
0.0816597509 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001225_030 |
0.083477091 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 13 |
Hb_000061_490 |
0.0837881803 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 14 |
Hb_004410_070 |
0.0838653623 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 15 |
Hb_003866_080 |
0.0840576345 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 16 |
Hb_005542_050 |
0.0841659829 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 17 |
Hb_012305_030 |
0.0847309518 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 18 |
Hb_001767_140 |
0.0849109525 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 19 |
Hb_002272_020 |
0.0850322863 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 20 |
Hb_002085_030 |
0.0863639917 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |