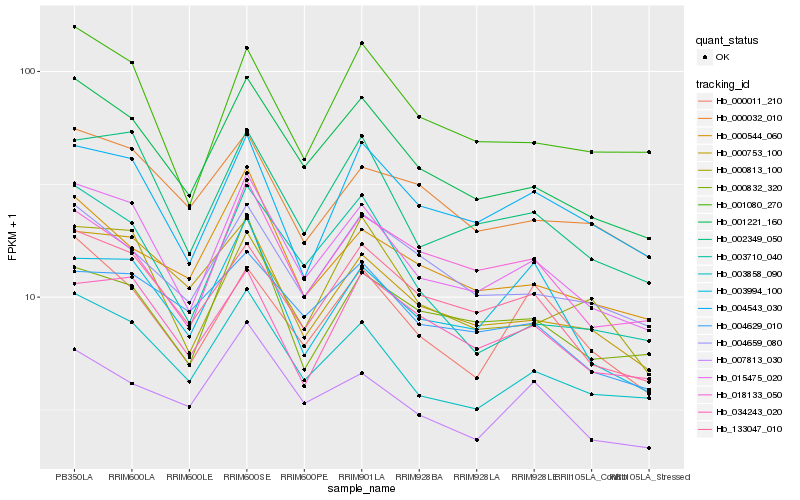
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015475_020 |
0.0 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 2 |
Hb_001221_160 |
0.0513409128 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 3 |
Hb_002349_050 |
0.0573026304 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 4 |
Hb_003858_090 |
0.0609441614 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 5 |
Hb_133047_010 |
0.0748555819 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_001080_270 |
0.0833448663 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 7 |
Hb_000753_100 |
0.087135529 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004543_030 |
0.0875466935 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 9 |
Hb_004659_080 |
0.0890105114 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 10 |
Hb_018133_050 |
0.0913733555 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 11 |
Hb_000813_100 |
0.0916176695 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000544_060 |
0.0933455026 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 13 |
Hb_034243_020 |
0.0938710703 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000011_210 |
0.0946911209 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 15 |
Hb_003710_040 |
0.0950650998 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 16 |
Hb_007813_030 |
0.0955201713 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 17 |
Hb_000032_010 |
0.0958126381 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 18 |
Hb_003994_100 |
0.0967063195 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004629_010 |
0.0969344501 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 20 |
Hb_000832_320 |
0.0971843224 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |