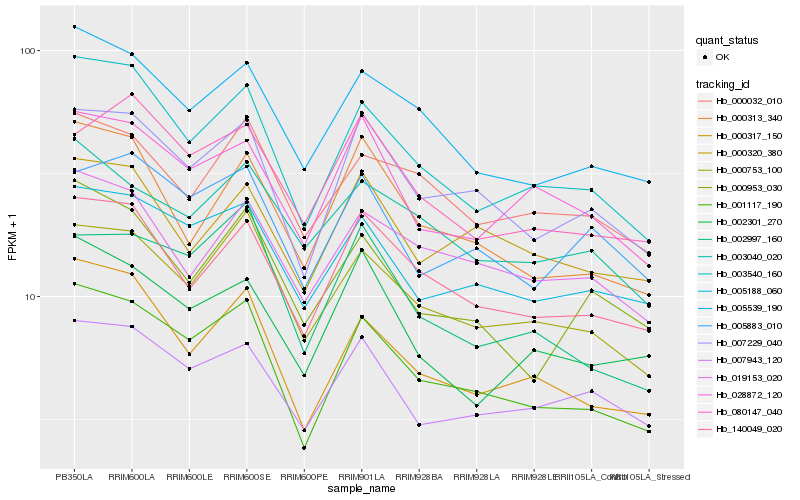
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_190 |
0.0 |
transcription factor |
TF Family: FAR1 |
FAR1-related sequence 11 [Theobroma cacao] |
| 2 |
Hb_003540_160 |
0.0566820786 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 3 |
Hb_007229_040 |
0.0616733438 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 4 |
Hb_000317_150 |
0.0633321318 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_000753_100 |
0.0736294854 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005539_190 |
0.0776788693 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 7 |
Hb_005188_060 |
0.0787569592 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 8 |
Hb_140049_020 |
0.0807992188 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 9 |
Hb_002997_160 |
0.0873113372 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007943_120 |
0.0911005962 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 11 |
Hb_000313_340 |
0.0919955487 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 12 |
Hb_028872_120 |
0.0981346208 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 13 |
Hb_000953_030 |
0.0987385201 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_005883_010 |
0.1001036273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_019153_020 |
0.1013854883 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000032_010 |
0.1014705704 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 17 |
Hb_002301_270 |
0.1035409602 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 18 |
Hb_080147_040 |
0.103596795 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 19 |
Hb_003040_020 |
0.1042829882 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 20 |
Hb_000320_380 |
0.104932666 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |