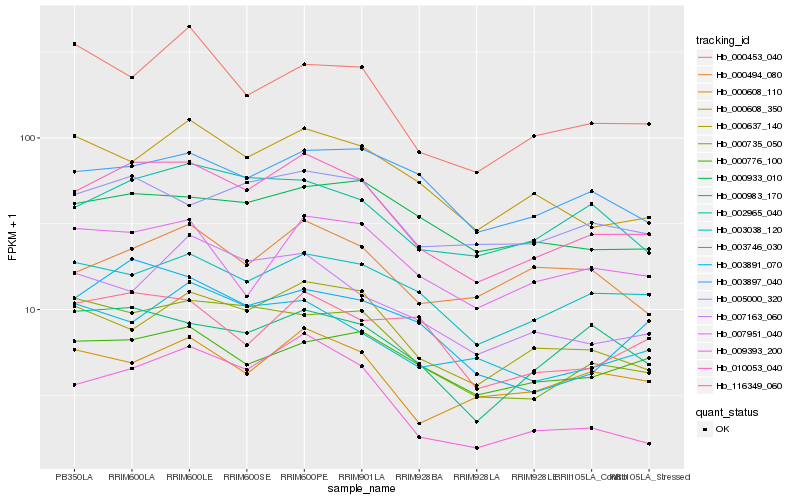
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010053_040 |
0.0 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 2 |
Hb_002965_040 |
0.1045735415 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 3 |
Hb_000735_050 |
0.1047466889 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 4 |
Hb_005000_320 |
0.1067135042 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |
| 5 |
Hb_000637_140 |
0.1070809688 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 6 |
Hb_003038_120 |
0.1098088962 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 7 |
Hb_003891_070 |
0.110829691 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 8 |
Hb_000608_350 |
0.1129088323 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 9 |
Hb_000983_170 |
0.114790773 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000608_110 |
0.1154015232 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 11 |
Hb_009393_200 |
0.1167527759 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 12 |
Hb_003746_030 |
0.1185452583 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 13 |
Hb_007951_040 |
0.1202454802 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 14 |
Hb_000933_010 |
0.1204908068 |
- |
- |
- |
| 15 |
Hb_116349_060 |
0.1208939582 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Brassica rapa] |
| 16 |
Hb_007163_060 |
0.1214688687 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 17 |
Hb_003897_040 |
0.1223744603 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 18 |
Hb_000494_080 |
0.1227850342 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000453_040 |
0.123409623 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 20 |
Hb_000776_100 |
0.1238501036 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |