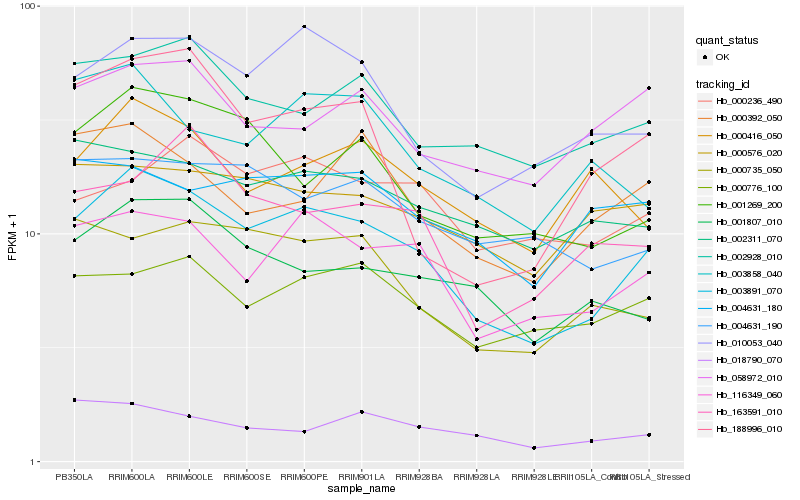
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003891_070 |
0.0 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 2 |
Hb_116349_060 |
0.0996010071 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Brassica rapa] |
| 3 |
Hb_010053_040 |
0.110829691 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 4 |
Hb_001269_200 |
0.1260167299 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 5 |
Hb_001807_010 |
0.1262324561 |
- |
- |
hypothetical protein MTR_6g077630 [Medicago truncatula] |
| 6 |
Hb_002928_010 |
0.1273821137 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 7 |
Hb_000735_050 |
0.1314902081 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 8 |
Hb_002311_070 |
0.1330554971 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 9 |
Hb_000776_100 |
0.1335439886 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 10 |
Hb_000576_020 |
0.1338727247 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 11 |
Hb_004631_190 |
0.1344204058 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 12 |
Hb_000416_050 |
0.1360473655 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 2 protein, putative [Ricinus communis] |
| 13 |
Hb_000392_050 |
0.1377325445 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 14 |
Hb_018790_070 |
0.1378836635 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 15 |
Hb_163591_010 |
0.1382356562 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |
| 16 |
Hb_003858_040 |
0.1391748718 |
- |
- |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 17 |
Hb_058972_010 |
0.1392166102 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000236_490 |
0.139788643 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 19 |
Hb_188996_010 |
0.1405125999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004631_180 |
0.1414741544 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |