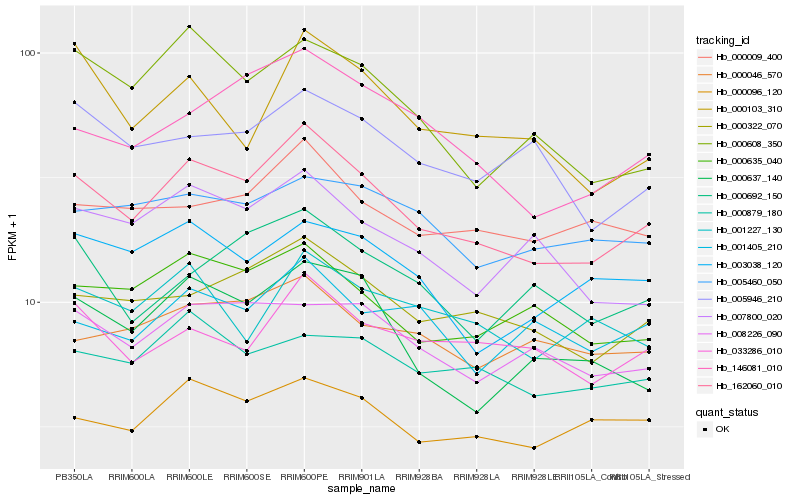
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162060_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000322_070 |
0.0784861909 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_146081_010 |
0.08637873 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 4 |
Hb_000635_040 |
0.0874831364 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 5 |
Hb_000096_120 |
0.0890678975 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 6 |
Hb_008226_090 |
0.093521538 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000009_400 |
0.0941216943 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 8 |
Hb_000637_140 |
0.0954487303 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 9 |
Hb_007800_020 |
0.095951895 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_033286_010 |
0.0968452562 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 11 |
Hb_003038_120 |
0.0980432411 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 12 |
Hb_000608_350 |
0.0980563984 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 13 |
Hb_000692_150 |
0.0989078874 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 3 [Jatropha curcas] |
| 14 |
Hb_000879_180 |
0.0992200833 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000046_570 |
0.0994633629 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 16 |
Hb_005460_050 |
0.1008184299 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 17 |
Hb_005946_210 |
0.1014514793 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 18 |
Hb_000103_310 |
0.1023595413 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 19 |
Hb_001405_210 |
0.1028622608 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 20 |
Hb_001227_130 |
0.1030241293 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |