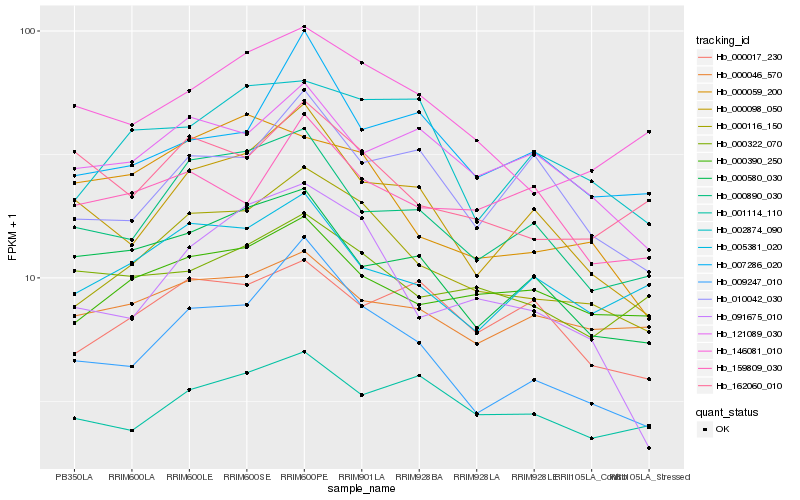
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_150 |
0.0 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 2 |
Hb_009247_010 |
0.0880774132 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_146081_010 |
0.1035077129 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 4 |
Hb_000890_030 |
0.1043193335 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 5 |
Hb_000390_250 |
0.105327564 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 6 |
Hb_091675_010 |
0.1123269335 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X1 [Jatropha curcas] |
| 7 |
Hb_000098_050 |
0.1131685942 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 8 |
Hb_002874_090 |
0.1138414852 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 9 |
Hb_005381_020 |
0.1150774959 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 10 |
Hb_121089_030 |
0.1213501186 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 11 |
Hb_010042_030 |
0.1240235554 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 12 |
Hb_000322_070 |
0.124236578 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_000580_030 |
0.1244407268 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 14 |
Hb_000046_570 |
0.1246782242 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 15 |
Hb_162060_010 |
0.1249044447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007286_020 |
0.1255086089 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 17 |
Hb_001114_110 |
0.1259656698 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 18 |
Hb_159809_030 |
0.1262442123 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000059_200 |
0.1266442427 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 20 |
Hb_000017_230 |
0.1267048006 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |