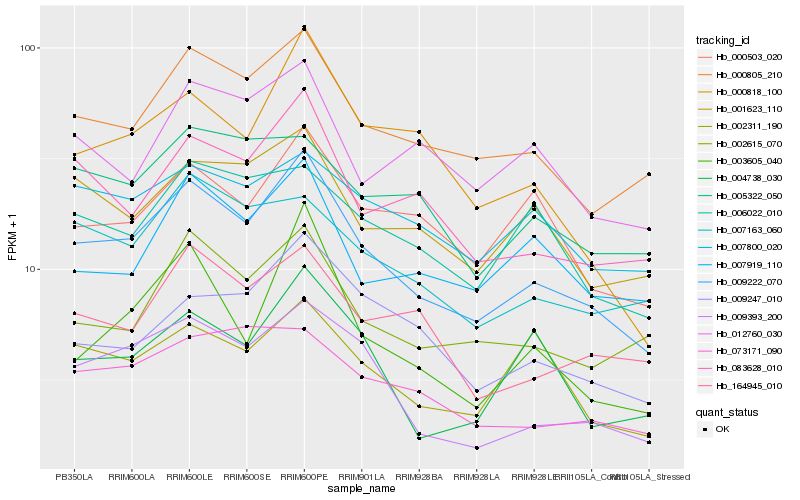
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009222_070 |
0.0 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 2 |
Hb_000805_210 |
0.08996441 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 3 |
Hb_009393_200 |
0.1049720898 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 4 |
Hb_083628_010 |
0.1058565334 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 5 |
Hb_002615_070 |
0.1121801864 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001623_110 |
0.1131943959 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 7 |
Hb_009247_010 |
0.1173115544 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000503_020 |
0.1194352224 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 9 |
Hb_164945_010 |
0.1196088451 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 10 |
Hb_007163_060 |
0.1202177726 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 11 |
Hb_007919_110 |
0.1215911943 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 12 |
Hb_000818_100 |
0.1220772754 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 13 |
Hb_073171_090 |
0.1234321376 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 14 |
Hb_002311_190 |
0.1271307486 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 15 |
Hb_006022_010 |
0.1277051385 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 16 |
Hb_007800_020 |
0.1285441033 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_012760_030 |
0.1286927203 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 18 |
Hb_005322_050 |
0.1291426374 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 19 |
Hb_003605_040 |
0.1299105238 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004738_030 |
0.131532749 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |