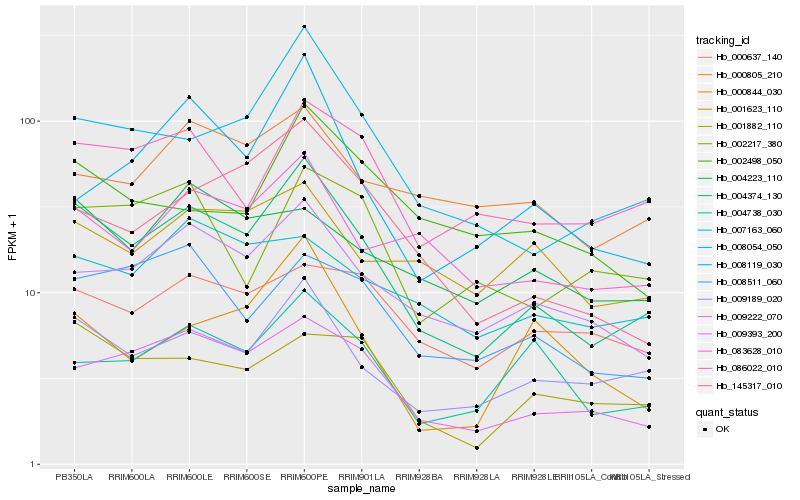
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004374_130 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 2 |
Hb_009189_020 |
0.1200407371 |
- |
- |
PREDICTED: CASP-like protein 4B4 [Jatropha curcas] |
| 3 |
Hb_009393_200 |
0.1348576521 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 4 |
Hb_008054_050 |
0.1381849797 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 5 |
Hb_009222_070 |
0.1419687232 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 6 |
Hb_145317_010 |
0.1549146291 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 7 |
Hb_083628_010 |
0.1572522335 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 8 |
Hb_086022_010 |
0.1603007401 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 9 |
Hb_004738_030 |
0.1693224142 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 10 |
Hb_001882_110 |
0.1714569793 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008511_060 |
0.1719783773 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 12 |
Hb_000844_030 |
0.1802147498 |
- |
- |
ent-kaurene oxidase, putative [Ricinus communis] |
| 13 |
Hb_000805_210 |
0.1811073335 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 14 |
Hb_008119_030 |
0.1811137777 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 15 |
Hb_002217_380 |
0.1818447451 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 16 |
Hb_002498_050 |
0.1819495942 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 17 |
Hb_001623_110 |
0.1823595431 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_000637_140 |
0.1853185924 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 19 |
Hb_007163_060 |
0.185626676 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 20 |
Hb_004223_110 |
0.1869873004 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |