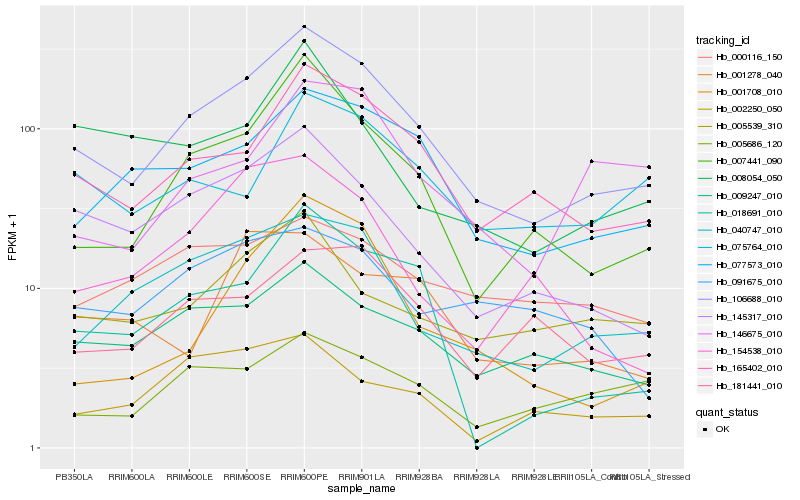
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106688_010 |
0.0 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 2 |
Hb_165402_010 |
0.1167967038 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 3 |
Hb_007441_090 |
0.125052808 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 4 |
Hb_077573_010 |
0.1375796364 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_040747_010 |
0.1403069323 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 6 |
Hb_145317_010 |
0.140578766 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 7 |
Hb_009247_010 |
0.1492783567 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_181441_010 |
0.1682548184 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 9 |
Hb_005539_310 |
0.1687452988 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 10 |
Hb_018691_010 |
0.1757711407 |
- |
- |
- |
| 11 |
Hb_154538_010 |
0.1760204922 |
- |
- |
- |
| 12 |
Hb_075764_010 |
0.1780404841 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 13 |
Hb_005686_120 |
0.1787343884 |
- |
- |
- |
| 14 |
Hb_001708_010 |
0.1826517503 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Malus domestica] |
| 15 |
Hb_146675_010 |
0.1837539009 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier-like isoform X2 [Malus domestica] |
| 16 |
Hb_008054_050 |
0.1844901716 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 17 |
Hb_091675_010 |
0.1861290968 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X1 [Jatropha curcas] |
| 18 |
Hb_002250_050 |
0.1862555177 |
- |
- |
PREDICTED: uncharacterized protein LOC105638972 [Jatropha curcas] |
| 19 |
Hb_001278_040 |
0.1880413822 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000116_150 |
0.1901956337 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |