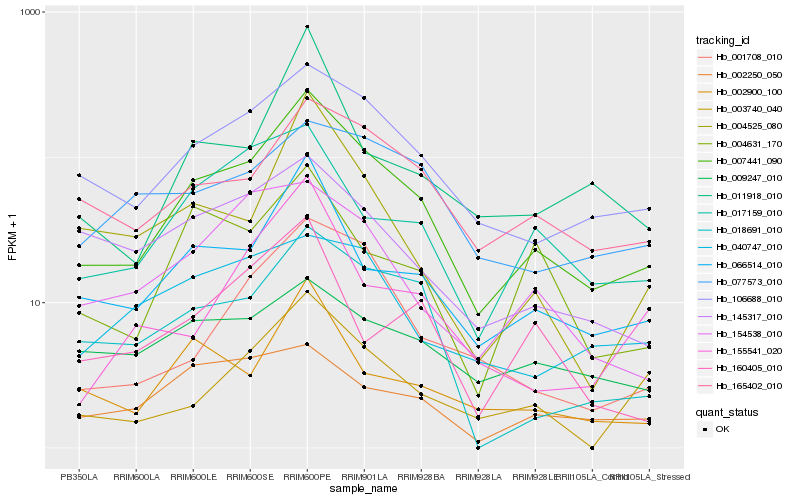
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_090 |
0.0 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 2 |
Hb_106688_010 |
0.125052808 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 3 |
Hb_066514_010 |
0.1408405521 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 4 |
Hb_165402_010 |
0.1582461335 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 5 |
Hb_018691_010 |
0.1693425841 |
- |
- |
- |
| 6 |
Hb_017159_010 |
0.1734409674 |
- |
- |
- |
| 7 |
Hb_001708_010 |
0.1776701434 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Malus domestica] |
| 8 |
Hb_002900_100 |
0.1781128805 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 9 |
Hb_154538_010 |
0.1815501463 |
- |
- |
- |
| 10 |
Hb_011918_010 |
0.1840301502 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 11 |
Hb_145317_010 |
0.1850889065 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 12 |
Hb_004525_080 |
0.1855784704 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 13 |
Hb_004631_170 |
0.1891876812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003740_040 |
0.1898469253 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase isozyme L3, putative [Ricinus communis] |
| 15 |
Hb_077573_010 |
0.1969322387 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_002250_050 |
0.1975314868 |
- |
- |
PREDICTED: uncharacterized protein LOC105638972 [Jatropha curcas] |
| 17 |
Hb_155541_020 |
0.2009522241 |
- |
- |
- |
| 18 |
Hb_160405_010 |
0.2013392576 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 19 |
Hb_040747_010 |
0.2017183055 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 20 |
Hb_009247_010 |
0.2030709857 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |