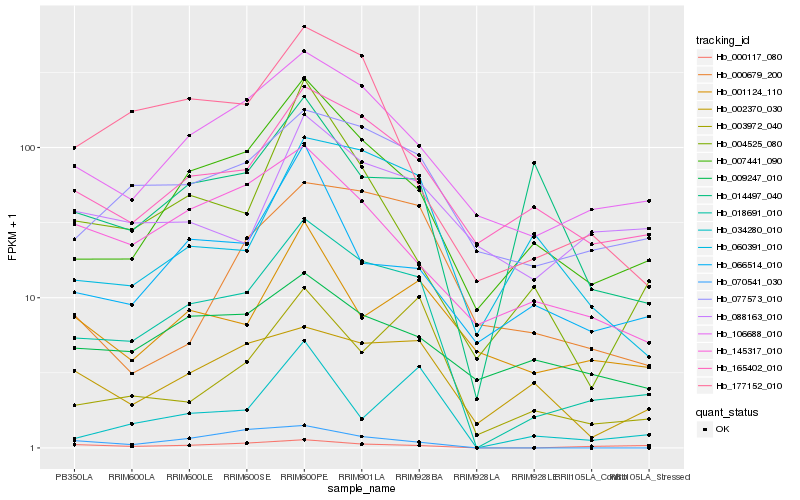
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018691_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_007441_090 |
0.1693425841 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 3 |
Hb_106688_010 |
0.1757711407 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 4 |
Hb_077573_010 |
0.1842170388 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_165402_010 |
0.1909055803 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 6 |
Hb_177152_010 |
0.205426816 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 7 |
Hb_145317_010 |
0.2168029846 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 8 |
Hb_003972_040 |
0.2182221027 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 9 |
Hb_060391_010 |
0.2187803514 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_034280_010 |
0.2244903262 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 11 |
Hb_070541_030 |
0.2309737859 |
- |
- |
- |
| 12 |
Hb_001124_110 |
0.2310752271 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_066514_010 |
0.2327020634 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 14 |
Hb_004525_080 |
0.2328196915 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 15 |
Hb_002370_030 |
0.2345134349 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 16 |
Hb_000679_200 |
0.2359140744 |
- |
- |
EIN3-binding F box protein 1 [Theobroma cacao] |
| 17 |
Hb_000117_080 |
0.2364446709 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 18 |
Hb_009247_010 |
0.2433105004 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_014497_040 |
0.2445099989 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_088163_010 |
0.2465552643 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |