| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
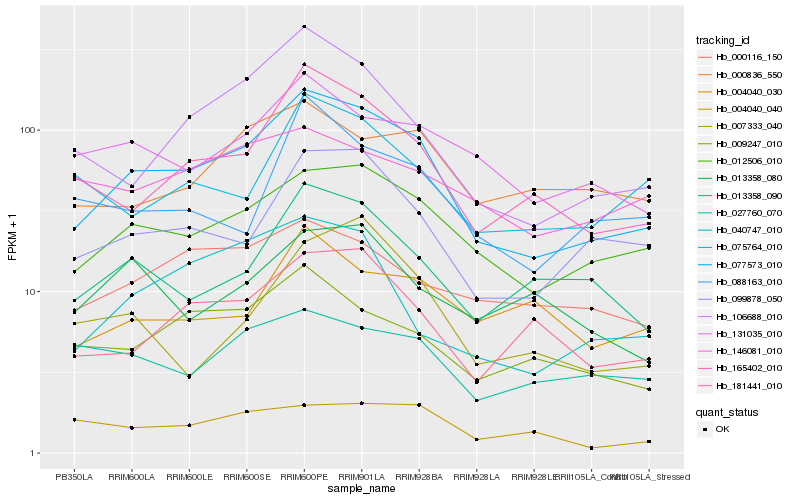
Hb_077573_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_012506_010 |
0.1132964895 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 3 |
Hb_165402_010 |
0.1296976668 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 4 |
Hb_106688_010 |
0.1375796364 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 5 |
Hb_099878_050 |
0.1376162905 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 6 |
Hb_181441_010 |
0.1478875491 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 7 |
Hb_009247_010 |
0.1528103595 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_013358_090 |
0.1529742823 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 9 |
Hb_131035_010 |
0.1546734483 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_040747_010 |
0.1615063386 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 11 |
Hb_000116_150 |
0.1638978534 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 12 |
Hb_000836_550 |
0.1662340504 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 13 |
Hb_075764_010 |
0.1697747387 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 14 |
Hb_088163_010 |
0.1710597192 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 15 |
Hb_027760_070 |
0.1770364838 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 16 |
Hb_004040_030 |
0.1779424046 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004040_040 |
0.1785470633 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 18 |
Hb_013358_080 |
0.1790882934 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 19 |
Hb_146081_010 |
0.181327049 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 20 |
Hb_007333_040 |
0.1839478121 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |