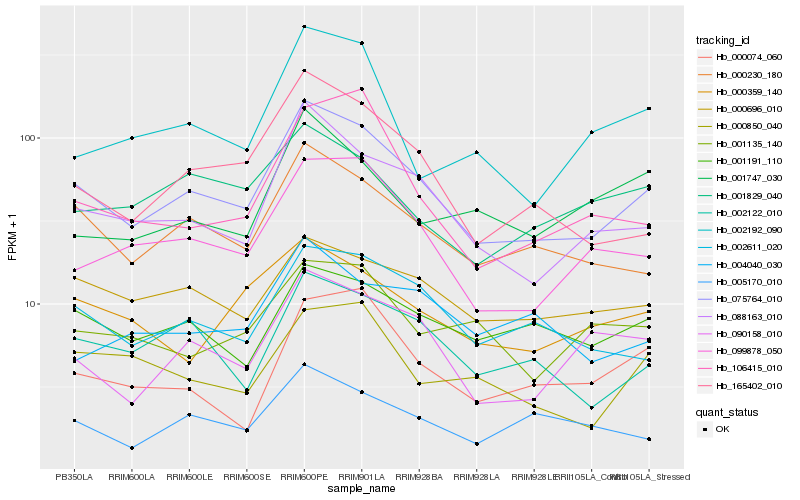
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075764_010 |
0.0 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 2 |
Hb_088163_010 |
0.0995900904 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 3 |
Hb_000230_180 |
0.1000461489 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 4 |
Hb_099878_050 |
0.1156419948 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 5 |
Hb_002611_020 |
0.1172121812 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 6 |
Hb_165402_010 |
0.1176477315 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 7 |
Hb_002122_010 |
0.1207609911 |
- |
- |
- |
| 8 |
Hb_090158_010 |
0.1269806385 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |
| 9 |
Hb_000696_010 |
0.1290510799 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 10 |
Hb_001191_110 |
0.1322923035 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 11 |
Hb_005170_010 |
0.1393910189 |
- |
- |
- |
| 12 |
Hb_000359_140 |
0.1411161018 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 13 |
Hb_001829_040 |
0.1417220147 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 14 |
Hb_002192_090 |
0.1441740374 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 15 |
Hb_001135_140 |
0.1460630764 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 16 |
Hb_106415_010 |
0.1465545318 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 17 |
Hb_000074_060 |
0.1471331409 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 18 |
Hb_000850_040 |
0.1478290708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001747_030 |
0.1524792812 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 20 |
Hb_004040_030 |
0.1529992932 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |